Principal Contact
Abstract:
Haemolysin (HlyE) is anessential virulence factor of Salmonella, Escherichia coli and other enteric bacteria.Although, a substantial degree of haemolytic activity is not seen under normal culture conditionsin these organisms, however, the non-haemolytic E. coli K-12 showed significant haemolytic activity under stress conditions. To confirm this phenomenon in other enteric bacteria, in this study, theproduction of haemolysin in Salmonella entericaserovar Typhi under stress conditions, likeoxygen and glucose starvationsin vitrowas investigated during March-December 2015.For this, S. typhi was cultured under oxygen or glucose starvation conditionseparately and this organismshowed high haemolytic activity.The activity was found to be much higher when both the conditions were applied together.Also, the role of the transcription factor SlyA of S. typhi was investigated on induction of haemolytic activity. When E. coli K-12 was transformed with plasmid containing the gene of SlyA,the recombinant bacteriawithout any starvation condition,also showed similar hemolytic activitythat was exhibited by S. typhi grown under oxygen and glucose starvation conditions. All these findings suggest that both environmental factors like oxygen or glucose starvation and overexpression of the transcription factor SlyAhave important role in inducing hlyE gene expression in S. typhi.
Key words: Glucose starvation; Haemolytic activity; hlyE gene; Oxygen starvation; Salmonella typhi
Key words: Glucose starvation; Haemolytic activity; hlyE gene; Oxygen starvation; Salmonella typhi
Introduction:
Salmonellae are the causative agents of a variety of diseases varying from local infections of the intestinal tract to systemic forms like typhoid fever.1 Typhoid fever is a fatalillness caused by Salmonella entericaserovar Typhi (S. typhi). Worldwide, about21.7 million cases ofillnesses and 2In,17,000 cases of deaths caused by typhoid fever are reported annually.2 South-central Asia and South-east Asia are the regions with high prevalence of typhoid fever (>100/100,000 cases/year).3 The disease is much more severe in Bangladesh, especially in young childrenwhere the S. typhi infectionis substantially higher.4 The proteomic analysis showed that the crucial steps of pathogenesis of S. typhi are a result of the production of a particular toxin, the haemolysin,a product of hlyE gene.5 Like many otherpore-formingtoxins, the HlyE toxin is also an important virulence factor among bacteria belonging to the Enterobacteriaceae.6,7 The HlyE, also denoted as ClyA and SheA, belonging to the family cytolysins, forms large, stable pores in target membranes.8 This toxin also causes haemolysis of erythrocytes and has apoptogenic effects on human and murine monocytes/macrophages.9,10 It has been reported that genes coding for close homologues of haemolysin are present in the S.entericaserovarTyphi or serovarParatyphi A and Shigellaflexnerigenomes.This haemolysin is also required for survival of thebacteria within the host macrophage.11,12 Furthermore, the wild‐type S. typhi and S. paratyphiA strains contain functional HlyE proteins, suggesting that the HlyE protein plays important roles in the pathogenesis of these organisms.8 Interestingly, the hlyEgene encoding this potentially toxin protein haemolysinis also present in the non-pathogenic Escherichia coli K-12 strain, which, however,normally exhibits no haemolytic activity under standard culture conditions.13 This could be due to the fact thatseveral other conditions may influence the regulation of hlyEgene expression in E. coli K‐12.12 In addition to the above findings, the non-haemolytic E. coli K-12, on the other hand,showed significant hemolytic activity under stress conditions like oxygen and glucose starvation,indicating that stress conditions confer haemolytic phenotype properties upon the non-haemolytic E. coli K-12.14 In the same study, an underlying molecular mechanism responsible for inducing hlyE gene expression in response to glucose starvation and anaerobic conditions in E. coli K-12 was also investigated by genetic analyses. Again,the hlyE gene hasalso been found to be present in S. typhii>.15 All the previous findings encouraged the authorsto confirm if the “stress” phenomenon is also applicable to other enteric bacteria like S. typhi. Therefore, in this study, the haemolytic activity of S. typhi under environmental stress conditions, like glucose and oxygen starvation, separately and together was investigated.Furthermore,conditions other than stress that induced haemolytic activity were investigated and it was found that recombinant E. coli,expressing the gene for transcription factor SlyA (Salmolysin)exhibited high hemolytic activity when cultured in normal conditions without any stresses.
Salmonellae are the causative agents of a variety of diseases varying from local infections of the intestinal tract to systemic forms like typhoid fever.1 Typhoid fever is a fatalillness caused by Salmonella entericaserovar Typhi (S. typhi). Worldwide, about21.7 million cases ofillnesses and 2In,17,000 cases of deaths caused by typhoid fever are reported annually.2 South-central Asia and South-east Asia are the regions with high prevalence of typhoid fever (>100/100,000 cases/year).3 The disease is much more severe in Bangladesh, especially in young childrenwhere the S. typhi infectionis substantially higher.4 The proteomic analysis showed that the crucial steps of pathogenesis of S. typhi are a result of the production of a particular toxin, the haemolysin,a product of hlyE gene.5 Like many otherpore-formingtoxins, the HlyE toxin is also an important virulence factor among bacteria belonging to the Enterobacteriaceae.6,7 The HlyE, also denoted as ClyA and SheA, belonging to the family cytolysins, forms large, stable pores in target membranes.8 This toxin also causes haemolysis of erythrocytes and has apoptogenic effects on human and murine monocytes/macrophages.9,10 It has been reported that genes coding for close homologues of haemolysin are present in the S.entericaserovarTyphi or serovarParatyphi A and Shigellaflexnerigenomes.This haemolysin is also required for survival of thebacteria within the host macrophage.11,12 Furthermore, the wild‐type S. typhi and S. paratyphiA strains contain functional HlyE proteins, suggesting that the HlyE protein plays important roles in the pathogenesis of these organisms.8 Interestingly, the hlyEgene encoding this potentially toxin protein haemolysinis also present in the non-pathogenic Escherichia coli K-12 strain, which, however,normally exhibits no haemolytic activity under standard culture conditions.13 This could be due to the fact thatseveral other conditions may influence the regulation of hlyEgene expression in E. coli K‐12.12 In addition to the above findings, the non-haemolytic E. coli K-12, on the other hand,showed significant hemolytic activity under stress conditions like oxygen and glucose starvation,indicating that stress conditions confer haemolytic phenotype properties upon the non-haemolytic E. coli K-12.14 In the same study, an underlying molecular mechanism responsible for inducing hlyE gene expression in response to glucose starvation and anaerobic conditions in E. coli K-12 was also investigated by genetic analyses. Again,the hlyE gene hasalso been found to be present in S. typhii>.15 All the previous findings encouraged the authorsto confirm if the “stress” phenomenon is also applicable to other enteric bacteria like S. typhi. Therefore, in this study, the haemolytic activity of S. typhi under environmental stress conditions, like glucose and oxygen starvation, separately and together was investigated.Furthermore,conditions other than stress that induced haemolytic activity were investigated and it was found that recombinant E. coli,expressing the gene for transcription factor SlyA (Salmolysin)exhibited high hemolytic activity when cultured in normal conditions without any stresses.
Materials and Methods:
Bacterial strains: All the organisms used in this study were obtained from the stock culture of the Department of Microbiology, University of Dhaka.The study was conducted during the period of March-December 2015.
Animal: New Zealand white rabbits (2-2.5 Kg body weight) were maintained in the Department of Microbiology, University of Dhaka and all experiments using animals were undertaken following ethical issues set by the Faculty of Biological Sciences, University of Dhaka.
Homology analysis of haemolysinof S. typhi and E. coli: Haemolysin sequence of S. typhi(NP_805266.1) and E. coli (AP_001807.1) were obtained from the protein sequence database Genpept (www.ncbi.nlm.nih.gov)and the homology between their amino acid sequences was determined using bioinformatics software tool ClustalW2.
Homology analysis of crp and fnr genes of S. typhi and E. coli: Sequences of the gene scrp [NC_003198.1 (4213325..4213957)] and fnr [NC_003198.1 (1355387..1356181)] of S. typhi and the sequences of the genes crp [NC_000913.3 (3486120..3486752)]and fnr [NC_000913.3 (1398774..1399526)] of E. coli were obtained from the Gene sequence database (www.ncbi.nlm.nih.gov). The homology analyses of the genes were performed using the bioinformatics software BLASTn.
Culturing S. typhi and E. coli under different conditions: To observe the haemolytic activity, the S. typhior E. coliK-12 strains were grown in both normal condition orunder different stress conditions. For normal culture condition, organisms were grown in Luria broth containing 0.2% glucose and incubated aerobically for 24 hours at 37oC. The stress conditions were appliedby oxygen or glucose starvation, either separately or together.For oxygen starvation condition, both organisms were grown in Luria broth with glucose supplement (0.2%) in sealed bottles and incubated in an anaerobic condition. On the other hand, for the glucose starvation condition, both organisms were grown aerobically but without any glucose supplement. Again, for both glucose and oxygen starvation conditions, both of these organisms were cultured without any glucose supplement in sealed bottlesand incubated anaerobically. All experiments were repeated twice to confirm the reproducibility of the results.
Protein extraction from bacteria: After 20 h of incubation of S. typhi or E. coli K-12 at normal or under different stress conditions, the antibiotic Polymyxin B was added to the media (5µg/ml) and cultures were further incubated for four more hours to break down the cells. Bacterial cultures were then centrifuged at 6,000 rpm for 10 min and the supernatants containing the bacterial proteins were collected.
Assay of haemolytic activity: The supernatants containing the bacterial proteins were analyzed for haemolytic activity,13 where 0.5ml of 1% rabbitred blood cell (RBC) solution was mixed with 4.5 ml of each of the bacterial proteins extracted fromeither S. typhi or E. coliK-12. The tubes werethen incubated for two hours at 37oC followed by centrifugation at 1,500 rpm for 10 min to pellet the erythrocytes. Absorbancesat 543nm were recorded spectrophotometrically to measure the amount of hemoglobin released into the supernatants.
Cloning of slyAgene: Isolation of genomic DNA, plasmid DNA and all DNA cloning procedures were carried out following the methods described by Sambrooket al.16 The gene slyA was amplified by Polymerase Chain Reaction (PCR) (Forward primer 5'-GCGTCAGACATGCATGCTTTAG-3'; Reverse primer 5'-GGTTACTGTCTGTCGACGCTAAACC-3').8 After restriction digestion of both purified PCR product and pACYC184 vector (Cmr) with SphI and SalI, the insertwas cloned into the dephosphorylated purified vector by cohesive end ligation reaction using DNA Blunting and Ligation Kit (K1512,Fermentas, UK). The successful ligation of slyA to the vector (pACYC184-slyAvector-insertconstruct) was confirmedby transformation into the chemo-competent E. coli DH5αcells. Molecular size of the vector-insert constructs isolated from the E. coli DH5α was confirmed by gel electrophoresis. Upon confirmation, pACYC184-slyA vector-insertconstructswere introduced into the chemo-competent E.coli K-12 cells and the transformed colonies were selected on chloramphenicol (12.5μg/ml)containing plates.
Statistical analysis: All data were statistically analyzed using ‘Student’s t-distribution’ to compare the differences in haemolytic activities of the extracted proteins at different culture and stress conditions.
Bacterial strains: All the organisms used in this study were obtained from the stock culture of the Department of Microbiology, University of Dhaka.The study was conducted during the period of March-December 2015.
Animal: New Zealand white rabbits (2-2.5 Kg body weight) were maintained in the Department of Microbiology, University of Dhaka and all experiments using animals were undertaken following ethical issues set by the Faculty of Biological Sciences, University of Dhaka.
Homology analysis of haemolysinof S. typhi and E. coli: Haemolysin sequence of S. typhi(NP_805266.1) and E. coli (AP_001807.1) were obtained from the protein sequence database Genpept (www.ncbi.nlm.nih.gov)and the homology between their amino acid sequences was determined using bioinformatics software tool ClustalW2.
Homology analysis of crp and fnr genes of S. typhi and E. coli: Sequences of the gene scrp [NC_003198.1 (4213325..4213957)] and fnr [NC_003198.1 (1355387..1356181)] of S. typhi and the sequences of the genes crp [NC_000913.3 (3486120..3486752)]and fnr [NC_000913.3 (1398774..1399526)] of E. coli were obtained from the Gene sequence database (www.ncbi.nlm.nih.gov). The homology analyses of the genes were performed using the bioinformatics software BLASTn.
Culturing S. typhi and E. coli under different conditions: To observe the haemolytic activity, the S. typhior E. coliK-12 strains were grown in both normal condition orunder different stress conditions. For normal culture condition, organisms were grown in Luria broth containing 0.2% glucose and incubated aerobically for 24 hours at 37oC. The stress conditions were appliedby oxygen or glucose starvation, either separately or together.For oxygen starvation condition, both organisms were grown in Luria broth with glucose supplement (0.2%) in sealed bottles and incubated in an anaerobic condition. On the other hand, for the glucose starvation condition, both organisms were grown aerobically but without any glucose supplement. Again, for both glucose and oxygen starvation conditions, both of these organisms were cultured without any glucose supplement in sealed bottlesand incubated anaerobically. All experiments were repeated twice to confirm the reproducibility of the results.
Protein extraction from bacteria: After 20 h of incubation of S. typhi or E. coli K-12 at normal or under different stress conditions, the antibiotic Polymyxin B was added to the media (5µg/ml) and cultures were further incubated for four more hours to break down the cells. Bacterial cultures were then centrifuged at 6,000 rpm for 10 min and the supernatants containing the bacterial proteins were collected.
Assay of haemolytic activity: The supernatants containing the bacterial proteins were analyzed for haemolytic activity,13 where 0.5ml of 1% rabbitred blood cell (RBC) solution was mixed with 4.5 ml of each of the bacterial proteins extracted fromeither S. typhi or E. coliK-12. The tubes werethen incubated for two hours at 37oC followed by centrifugation at 1,500 rpm for 10 min to pellet the erythrocytes. Absorbancesat 543nm were recorded spectrophotometrically to measure the amount of hemoglobin released into the supernatants.
Cloning of slyAgene: Isolation of genomic DNA, plasmid DNA and all DNA cloning procedures were carried out following the methods described by Sambrooket al.16 The gene slyA was amplified by Polymerase Chain Reaction (PCR) (Forward primer 5'-GCGTCAGACATGCATGCTTTAG-3'; Reverse primer 5'-GGTTACTGTCTGTCGACGCTAAACC-3').8 After restriction digestion of both purified PCR product and pACYC184 vector (Cmr) with SphI and SalI, the insertwas cloned into the dephosphorylated purified vector by cohesive end ligation reaction using DNA Blunting and Ligation Kit (K1512,Fermentas, UK). The successful ligation of slyA to the vector (pACYC184-slyAvector-insertconstruct) was confirmedby transformation into the chemo-competent E. coli DH5αcells. Molecular size of the vector-insert constructs isolated from the E. coli DH5α was confirmed by gel electrophoresis. Upon confirmation, pACYC184-slyA vector-insertconstructswere introduced into the chemo-competent E.coli K-12 cells and the transformed colonies were selected on chloramphenicol (12.5μg/ml)containing plates.
Statistical analysis: All data were statistically analyzed using ‘Student’s t-distribution’ to compare the differences in haemolytic activities of the extracted proteins at different culture and stress conditions.
Result:
Haemolytic activities of cell extracts of S. typhi grown under stress conditions like oxygen or glucose starvation, either separately or together, were found to be significantly higher when compared with the normal growth conditions(figure 1).Interestingly,the haemolytic activity was found to be higher in glucose starvation than in oxygen starvation(p<0.001). Again, the hemolytic activity exhibited by S. typhi was found to be the highest when both of these starvation conditions were applied together than any of the stresses applied alone (p<0.001).
Thestudy also showed that the E. coli K-12 grown under oxygen and glucose starvation conditions, either alone or together, had similarpattern of hemolytic activitieslike that of S. typhi grown under the same conditions(figure 2).This result also indicates that, under separate oxygen or glucose starvation conditions, S. typhi showed 1.51 folds and 1.11 folds more hemolytic activities,respectively, when compared with the hemolytic activities of E. coli K-12 under same conditions. Again, the magnitude of hemolysis was found to be highest in E. coli K-12, when both oxygen and glucose starvation were applied togther, and the value was comparable to that of the hemolytic activity shown by S. typhi grown underthe same conditions (figure 2). However, the haemolytic activity displayed by E. coli under oxygen starvation condition was 51% higher and under glucose starvation condition was 11.8% higher than that displayed by S.typhi under the same conditions. In addition to the stress factors, other conditions that might induce haemolytic activity in S. typhi were also investigated. As previously reported, the SlyA, a transcription factor that positively regulates haemolysin production, is present in both S.typhi and E. coli K-12.13,15 To assess the effect of overexpression of SlyA on induction of haemolytic activity in normal condition, i.e. without any stress, theslyA gene from S. typhi was isolatedand cloned into E. coli K-12. The transformed E. coli was then grownunder normal condition and the degree of haemolytic activity was observed. When the cell extract of transformed cellswas observed for the haemolytic activity, surprisingly a significant degree of haemolysis of RBC was found to occur, when compared with the cell extract of the non-transformed E. coli K-12 (figure 3).
indicated that these particular stresses may be important for turning on the hlyE gene responsible for hemolysin production in S. typhi that ultimately caused hemolysis. This result was in accordance with the findings of Westermerk et al14 where the non-hemolytic E. coli K-12 also showed significant hemolytic activity under stress conditions.
Again, the comparison of the haemolytic activities of S. typhi and E. coli (figure 2)
Figure.2: Comparison of haemolytic activities of cell extracts ofS. typhi with that of E. coli K-12 grown under normal and stress conditions like oxygen or glucose starvation, either separately or together. Under separate oxygen or glucose starvation conditions, S. typhi showed more hemolytic activities when compared with the hemolytic activities of E. coli K-12 under same conditions (p<0.001).
clearly indicated that the haemolytic activities induced in S. typhi under stress conditions is quite consistent to our predictions, since S. typhi also harbors hlyE gene like that of E. coli K-12. Though the hlyE gene is present in E. coli K-12, however, it is silent in normal culture condition and its expression can be induced through stress conditions. It has been reported that two members of the cAMP Receptor Protein (CRP) family of transcription factors control the expression of hlyE in E. coli K-12, where CRP enhances hlyE expression in response to glucose starvation and Fumarate Nitrate Reduction (FNR) regulatory protein enhances hlyE expression in response to oxygen starvation.15
In this context, using bioinformatics analysis, in this study it was observed that the genes crp and fnr of transcription factors CRP and FNR respectively, wereboth found in S. typhi and these two genes of S. typhi were 88% identical with the crp and fnr genes of E. coli K-12, respectively. It was also observed that both S. typhi and E. coli K-12 contain functional homologs of hlyE and the protein encoded by the S. typhi is 90% identical in amino acid sequence to that of the hlyE of E. coli K-12. In reality, the S. typhi does not express this gene in normal culture conditions like that of E. coli K-12.Therefore, it is evident from the current study that S. typhi has similar strict control mechanisms for the hlyE gene and expression of hlyE gene can be induced in S. typhi under oxygen and/or glucose starvation conditions that confer the hemolytic ability upon S. typhi. This novel finding clearly indicates that environmen-tal starvation conditions may act as stress factors for the induction of the hlyE gene to produce hemolysin in S. typhi.
Also, the increased haemolytic activity of E. coli K-12 transformed with the transcription factor SlyA gene from S. typhi (figure 3). clearly indicated that overproduction of SlyA in E. coli K-12 is independent of any culture condition. The result also suggests that the hemolytic activity can be induced in a different bacterial strain when the gene for transcription factor SlyA was cloned from another bacterial strain. This finding supports the phenomenon that SlyA overproduction antagonizes the negative effects of the regulatory protein H-NS, a nucleoid structuring protein that represses hlyE gene expression.15
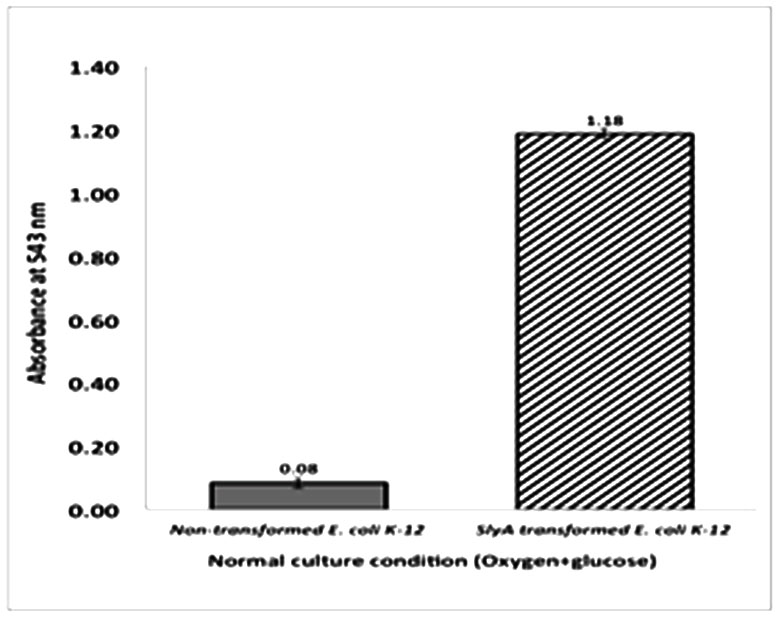
Based on this findings of molecular mechanisms, we assessed the degree of hemolysis seen in the hemolytic assay with RBC, after overexpression of slyA gene under normal culture conditions, and all results indicated that hlyE gene might be expressed in response to appropriate environmental signals.
Haemolytic activity can be considered to be one kind of virulence factor.17 Therefore, findings of this study are very significant in clinical perspectives, since stress conditions may induce virulent properties in bacterial strains. There is evidence that stresses like starvation, acidic pH and heat shock may induce the expression of some virulence genes.18 Again, it has been suggested that conditions stimulating the production of the haemolysin might be encountered during infection by ,i>E. coli strains.13 As both S. typhi and E. coli are enteric bacteria, therefore, it is assumed that these organisms may encounter oxygen starvation condition in intestinal environment which may induce the production of haemolysin in the host. This is again supported by the report that anaerobiosis has been shown to induce the invasion phenotype in Salmonella.19-21 Hence, during the course of their infection, these bacteria may encounter anaerobic condition or limited glucose, which may induce haemolysin production. Therefore, all our results along with reports of other investigators clearly indicate that, S. typhi may exhibit hemolytic activity under glucose or/and oxygen starvation conditions. Again, the SlyA transcription factor may also induce haemolysin production, if it is overexpressed in vivo, without going through any starvation conditions.
Conclusion
Findings of this study may also help to understand how Salmonella survive in the macrophages and how the haemolysin may help S. typhi in causing infections as a crucial virulence factor. Therefore, the haemolysin protein can be a potential antigenic target for development of a vaccine as an alternative therapeutic agent for Salmonella infections. Previously, in Bangladesh, studies on anti-HlyE responses in patients have been carried out with a view to developing improved diagnostic assays.22 In this novel study, it was assayed production of HlyE toxin under various conditions in order to analyse this protein with a view to developing a novel vaccine against this toxin in future in Bangladesh.
Acknowledgments: This project has been partially funded by University Grants Commission (UGC), Bangladesh.We would like to heartily thank Dr. Siraje Arif Mahmud for his expert assistance with this manuscript.
Haemolytic activities of cell extracts of S. typhi grown under stress conditions like oxygen or glucose starvation, either separately or together, were found to be significantly higher when compared with the normal growth conditions(figure 1).Interestingly,the haemolytic activity was found to be higher in glucose starvation than in oxygen starvation(p<0.001). Again, the hemolytic activity exhibited by S. typhi was found to be the highest when both of these starvation conditions were applied together than any of the stresses applied alone (p<0.001).
Thestudy also showed that the E. coli K-12 grown under oxygen and glucose starvation conditions, either alone or together, had similarpattern of hemolytic activitieslike that of S. typhi grown under the same conditions(figure 2).This result also indicates that, under separate oxygen or glucose starvation conditions, S. typhi showed 1.51 folds and 1.11 folds more hemolytic activities,respectively, when compared with the hemolytic activities of E. coli K-12 under same conditions. Again, the magnitude of hemolysis was found to be highest in E. coli K-12, when both oxygen and glucose starvation were applied togther, and the value was comparable to that of the hemolytic activity shown by S. typhi grown underthe same conditions (figure 2). However, the haemolytic activity displayed by E. coli under oxygen starvation condition was 51% higher and under glucose starvation condition was 11.8% higher than that displayed by S.typhi under the same conditions. In addition to the stress factors, other conditions that might induce haemolytic activity in S. typhi were also investigated. As previously reported, the SlyA, a transcription factor that positively regulates haemolysin production, is present in both S.typhi and E. coli K-12.13,15 To assess the effect of overexpression of SlyA on induction of haemolytic activity in normal condition, i.e. without any stress, theslyA gene from S. typhi was isolatedand cloned into E. coli K-12. The transformed E. coli was then grownunder normal condition and the degree of haemolytic activity was observed. When the cell extract of transformed cellswas observed for the haemolytic activity, surprisingly a significant degree of haemolysis of RBC was found to occur, when compared with the cell extract of the non-transformed E. coli K-12 (figure 3).
Discussion
The higher haemolytic activities showed by S. typhi at glucose and oxygen starvation conditions (figure.1).indicated that these particular stresses may be important for turning on the hlyE gene responsible for hemolysin production in S. typhi that ultimately caused hemolysis. This result was in accordance with the findings of Westermerk et al14 where the non-hemolytic E. coli K-12 also showed significant hemolytic activity under stress conditions.
Again, the comparison of the haemolytic activities of S. typhi and E. coli (figure 2)
clearly indicated that the haemolytic activities induced in S. typhi under stress conditions is quite consistent to our predictions, since S. typhi also harbors hlyE gene like that of E. coli K-12. Though the hlyE gene is present in E. coli K-12, however, it is silent in normal culture condition and its expression can be induced through stress conditions. It has been reported that two members of the cAMP Receptor Protein (CRP) family of transcription factors control the expression of hlyE in E. coli K-12, where CRP enhances hlyE expression in response to glucose starvation and Fumarate Nitrate Reduction (FNR) regulatory protein enhances hlyE expression in response to oxygen starvation.15
In this context, using bioinformatics analysis, in this study it was observed that the genes crp and fnr of transcription factors CRP and FNR respectively, wereboth found in S. typhi and these two genes of S. typhi were 88% identical with the crp and fnr genes of E. coli K-12, respectively. It was also observed that both S. typhi and E. coli K-12 contain functional homologs of hlyE and the protein encoded by the S. typhi is 90% identical in amino acid sequence to that of the hlyE of E. coli K-12. In reality, the S. typhi does not express this gene in normal culture conditions like that of E. coli K-12.Therefore, it is evident from the current study that S. typhi has similar strict control mechanisms for the hlyE gene and expression of hlyE gene can be induced in S. typhi under oxygen and/or glucose starvation conditions that confer the hemolytic ability upon S. typhi. This novel finding clearly indicates that environmen-tal starvation conditions may act as stress factors for the induction of the hlyE gene to produce hemolysin in S. typhi.
Also, the increased haemolytic activity of E. coli K-12 transformed with the transcription factor SlyA gene from S. typhi (figure 3). clearly indicated that overproduction of SlyA in E. coli K-12 is independent of any culture condition. The result also suggests that the hemolytic activity can be induced in a different bacterial strain when the gene for transcription factor SlyA was cloned from another bacterial strain. This finding supports the phenomenon that SlyA overproduction antagonizes the negative effects of the regulatory protein H-NS, a nucleoid structuring protein that represses hlyE gene expression.15

Based on this findings of molecular mechanisms, we assessed the degree of hemolysis seen in the hemolytic assay with RBC, after overexpression of slyA gene under normal culture conditions, and all results indicated that hlyE gene might be expressed in response to appropriate environmental signals.
Haemolytic activity can be considered to be one kind of virulence factor.17 Therefore, findings of this study are very significant in clinical perspectives, since stress conditions may induce virulent properties in bacterial strains. There is evidence that stresses like starvation, acidic pH and heat shock may induce the expression of some virulence genes.18 Again, it has been suggested that conditions stimulating the production of the haemolysin might be encountered during infection by ,i>E. coli strains.13 As both S. typhi and E. coli are enteric bacteria, therefore, it is assumed that these organisms may encounter oxygen starvation condition in intestinal environment which may induce the production of haemolysin in the host. This is again supported by the report that anaerobiosis has been shown to induce the invasion phenotype in Salmonella.19-21 Hence, during the course of their infection, these bacteria may encounter anaerobic condition or limited glucose, which may induce haemolysin production. Therefore, all our results along with reports of other investigators clearly indicate that, S. typhi may exhibit hemolytic activity under glucose or/and oxygen starvation conditions. Again, the SlyA transcription factor may also induce haemolysin production, if it is overexpressed in vivo, without going through any starvation conditions.
Conclusion
Findings of this study may also help to understand how Salmonella survive in the macrophages and how the haemolysin may help S. typhi in causing infections as a crucial virulence factor. Therefore, the haemolysin protein can be a potential antigenic target for development of a vaccine as an alternative therapeutic agent for Salmonella infections. Previously, in Bangladesh, studies on anti-HlyE responses in patients have been carried out with a view to developing improved diagnostic assays.22 In this novel study, it was assayed production of HlyE toxin under various conditions in order to analyse this protein with a view to developing a novel vaccine against this toxin in future in Bangladesh.
Acknowledgments: This project has been partially funded by University Grants Commission (UGC), Bangladesh.We would like to heartily thank Dr. Siraje Arif Mahmud for his expert assistance with this manuscript.
References
- Ludwig A, Tengel C, Bauer S, Bubert A, Benz R, Mollenkopf HJ, et al. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol Gen Genet.1995;249:474–86.
- Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis.2010;50(2):241-46.
- Crump JA, Luby SP, Mintz ED.The global burden of typhoid fever. Bulletin of World Health Organization.2004;82:346-53.
- Saha SK, Baqui AH, Hanif M, Darmstadt GL, Ruhulamin M, Nagatake T, et al. Typhoid fever in Bangladesh: Implications for vaccination policy. Pediatr Infect Dis J. 2001;20(5):521-24.
- Ansong C, Yoon H, Norbeck AD, Gustin JK, McDermott JE, Mottaz HM,et al.Proteomics analysis of the causative agent of typhoid fever. J Proteome Res.2008;7:546-57.
- Alouf JE. Pore-forming bacterial protein toxins: an overview. Curr Top Microbiol Immunol. 2001;257:1-14.
- Parker MW, Feil SC. Pore-forming protein toxins: from structure to function. Prog Biophys Mol Biol. 2005;88:91–142.
- vonRhein C, Hunfeld KP, Ludwig A. Serologic evidence for effective production of cytolysin A in Salmonella enterica serovars Typhi and Paratyphi A during human infection. Infect Immun.2006;74:6505-08.
- Lai XH, Arencibia I, Johansson A, Wai SN, Oscarsson J, Kalfas S,et al.Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect Immun. 2000;68:4363-67.
- Oscarsson J, Westermark M, Lofdahl S, Olsen B, Palmgren H, Mizunoe Y, et al. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars Typhi and Paratyphi A. Infect Immun. 2002;70(10):5759-69.
- Libby SJ, Goebel W, Ludwig A, Buchmeier N, Bowel F, Fang FC, et al. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci. 1994;91:489-93.
- Hunt S, Green J, Artymiuk PJ.Hemolysin E (HlyE, ClyA, SheA) and related toxins. In:Proteins: Membrane Binding and Pore Formation. V 677. Anderluh G, Lakey J editorss.New York, USA, Springer Science+Business Media, 2010,p116-26.
- Ludwig A, Bauer S, Benz R, Bergmann B, Goebel W. Analysis of the SlyA-controlled expression, sub-cellular localization and pore-forming activity of a 34kDa haemolysin (ClyA) from Escherichia coli K-12. Mol Microbiol. 1999;31:557–67.
- Westermark M, Oscarsson J, Mizunoe Y, Urbonavi-ciene J, Uhlin BE. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J Bacteriol.2000;182(22):6347-57.
- Wyborn NR, Stapleton MR, Norte VA, Roberts RE, Grafton J, Green J. Regulation of Escherichia coli Hemolysin E expression by H-NS and Salmonella SlyA. J Bacteriol. 2004;186(6):1620-28.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual: 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989.
- Goebel W, Chakraborty T, Kreft J. Bacterial hemolysins as virulence factors. Antonie van Leeuwenhoek.1988;54:453-63.
- Mekalanos JJ. Environmental signals controlling expression of virulence determinants in bacteria.J Bacteriol.1992;174(1):1-7.
- Ernst RK, Dombroski DM, Merrick JM. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014-16.
- Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci. 1990;87:4304-08.
- Schiemann DA, Shope SR. Anaerobic growth of Salmonella typhimuriumresults in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437-40.
- Charles RC, Sheikh A, Krastins B, Harris JB, Bhuiyan MS, LaRocque RC, et al. Characteriza-tion of anti-Salmonella enterica serotype Typhi antibody responses in bacteremic Bangladeshi patients by an immunoaffinity proteomics-based technology.Clin Vaccine Immunol. 2010; 17(8):1188-95.