Key words:Systemic lupus erythematosus, Major histocompatibility complex, human leukocyte antigen, polymerase chain reaction.
Health problem due to rheumatic disorders are increasing day by day. Now-a-days they are the commonest cause of morbidity. Among the mul-tisystem rheumatic disorders systemic lupus ery-thematosus (SLE) is the commonest autoimmune disorder.1 This systemic autoimmune disease af-fects multiple organs, which is characterized by production of autoantibodies against a variety of self antigens such as double stranded DNA (dsDNA), intracellular ribonucleoproteins and membrane phospholipids. Severity, acquisition risk and clinical manifestations of this disease may vary by ethnicity, geography and sex. It pri-marily affects female (female: male-9:1), espe-cially during their childbearing age.2 Some non-European populations such as African Ameri-cans, Hispanics and Asians are at increased risk for acquiring the disease. Although the exact aetiology of SLE remains vague, genetic predisposition, environmental and hormonal factors play important role in its pathogenesis. The high concordance rate for SLE in monozygotic twins than in dizygotic twins or siblings (24 -56% versus 2-5%) and the high sibling recurrence risk ratio of patients with SLE (between 8-fold and 29-fold higher than general population) proved a strong genetic contribution for development of the disease.3,4
SLE is a polygenic disorder. Effects of multiple genes are required for development of SLE. Al-though in rare cases SLE may be associated with deficiency of a single gene. The search for gene that predisposes a person to develop SLE has been going on through association studies of candidate genes and genome wide linkage analy-sis. A large number of genetic regions have been identified that may contain susceptibility gene. Among them, eight susceptibility loci have been confirmed: 1q23, 1q31, 1q41-42, 2q37, 4p16, 6p21, 12q24 and 16q12-13.5,6 But region 6p21.1- q15 and 20q11- q13.13 have reached the threshold level for significant linkage.6 Of the genetic elements, the genes of the major histocompatibility complex (MHC) have been most extensively studied for their contribution to development of SLE. MHC, located on chromosome 6p21.3, harbours a gene reach and transcriptionally active segment that encodes for immunologically important gene, including highly polymorphic human leukocyte antigen (HLA) class I and class II genes. Class I region contains HLA-A, -B, -C genes, which present antigenic peptides to CD8+ T cells. Class II region contains highly polymorphic HLA-DR, -DQ and -DP genes. They present antigenic peptides to CD4+ T cells. The class III region lies between class I and class II that contains many immune related genes, including cy-tokines, tumor necrosis factor-α, lymphotoxin-α, the complement components C2, C4 and Factor B.7
Association of Human leukocyte antigen (HLA) with SLE was first reported in 1971 and it was HLA class I B8.8 But later studies showed that association of HLA class II genes are more strong and consistent with SLE than HLA class I genes. Among the HLA class II, HLA-DR2 and DR3 alleles are proved to be associated with a two to five fold risk for the development of SLE.9 However this association varies considera-bly in different races and ethnic groups due to genetic heterogenecity, which is much more common among populations of SLE.10,11 Amongst Europeans most studies showed associ-ation of SLE with HLA-DR3 and in Asian coun-tries most studies found association with DR2. But data regarding association study of SLE with HLA among Bangladeshi is not available. In addition, several studies also suggested that the contribution of HLA class II genes in SLE is also involved in specific autoantibody production like HLA-DR2 and DR3 are associated with development of anti-dsDNA and anti-Sm autoantibodies.12 It has also been found that DR2/DR3 genotype may predispose to development of autoantibodies in unaffected family members of SLE cases.7 So, identification of candidate genes and understanding of genetic influences is necessary to comprehend the pathophysiology of this autoimmune disease and to predict high risk patients who carry genetic susceptibility factors in general population or within families. Therefore, the present study was undertaken to determine the association of HLA-DR (DR1 to DR10) with development of SLE in SLE cases.
This case control study was done during the pe-riod of March 2013 to February 2014. Samples were collected from SLE clinic, Department of Rheumatology, BSMMU, Dhaka. Laboratory works were performed in the Department of Mi-crobiology and Immunology, BSMMU, Dhaka. A total of 46 diagnosed case of SLE were enrolled in this study after taking informed writ-ten consent. All patients met at least 4 criteria out of the 11 of the 1997 update of the 1982 American College of Rheumatology revised criteria for diagnosis of SLE.13 A complete physical examination was performed and symptoms were noted. History of other associated autoimmune disease and past medical history were taken from previous records. Patients having other diagnosed autoimmune diseases in association with SLE were excludedfor the present study. A total 46 healthy volunteer age and sex matched unrelated healthy controls were enrolled in this study after taking informed written consent. The controls population were medical students and nurses. Control population included in this study were negative for anti-dsDNA, anti-Sm and did not have history of rheumatic disorders. Persons having family history of SLE and other rheumatic disorders were excluded as control population. Buccal swabs were collected from both cases and controls for HLA-DR typing.
HLA typing: Genomic DNA was extracted from buccal swab samples by using Chelex® 100 fol-lowed by protein digestion in proteinase K solu-tion.14 HLA-DR typing was done by using poly-merase chain reaction sequence specific primer (PCR-SSP) (MorganTM HLA SSP DRB typing kit) using low resolution typing method. The amplified DNA was examined by agar gel elec-trophoresis that separates the DNA fragments by size. Specific HLA-DRB type was determined using the worksheet (supplied along with the kit).
Statistical analysis: The allelic frequencies of HLA-DR in cases and controls were compared using chi-square with Yates correction (by using online available GraphPad QuickCalcs calcula-tor). The strength of association between HLA-DR antigens and SLE was estimated by relative risk (RR) and 95% confidence intervals (95% CI) using online MedCalc software (Version- 12.7.8.0). The relative risk was determined by the odd ratio (OR). p value of less than 0.05 was taken to be significant. p corrected (p corr) was determined by multiplying p value with the number of HLA alleles tested (Bonferroni's correction).
A total of 46 cases with SLE and 46 healthy controls were enrolled in this study. There were 44 (95.65%) female and 2 (4.35%) male in cases. Their mean age was 27.05 ± 8.17 years (mean ± SD), ranging from 12.5-45 years. In control group, there were 44 (95.65%) female and 2 (4.35%) male with a mean age of 26.91 ± 5.96 years (mean ± SD), ranging from 16-45 years. In both case and control group female: male ratio was 22: 1 in table I
Study population |
Sex |
Age (year) |
|||
Female |
Male |
Female: male |
Mea ± SD |
Age range |
|
Case |
44 (95.65) |
2 (4.35) |
22:1 |
27.05 ± 8.17 |
12.5 - 45 |
Control |
44 (95.65) |
2 (4.35) |
22:1 |
26.91 ± 5.96 |
16 - 45 |
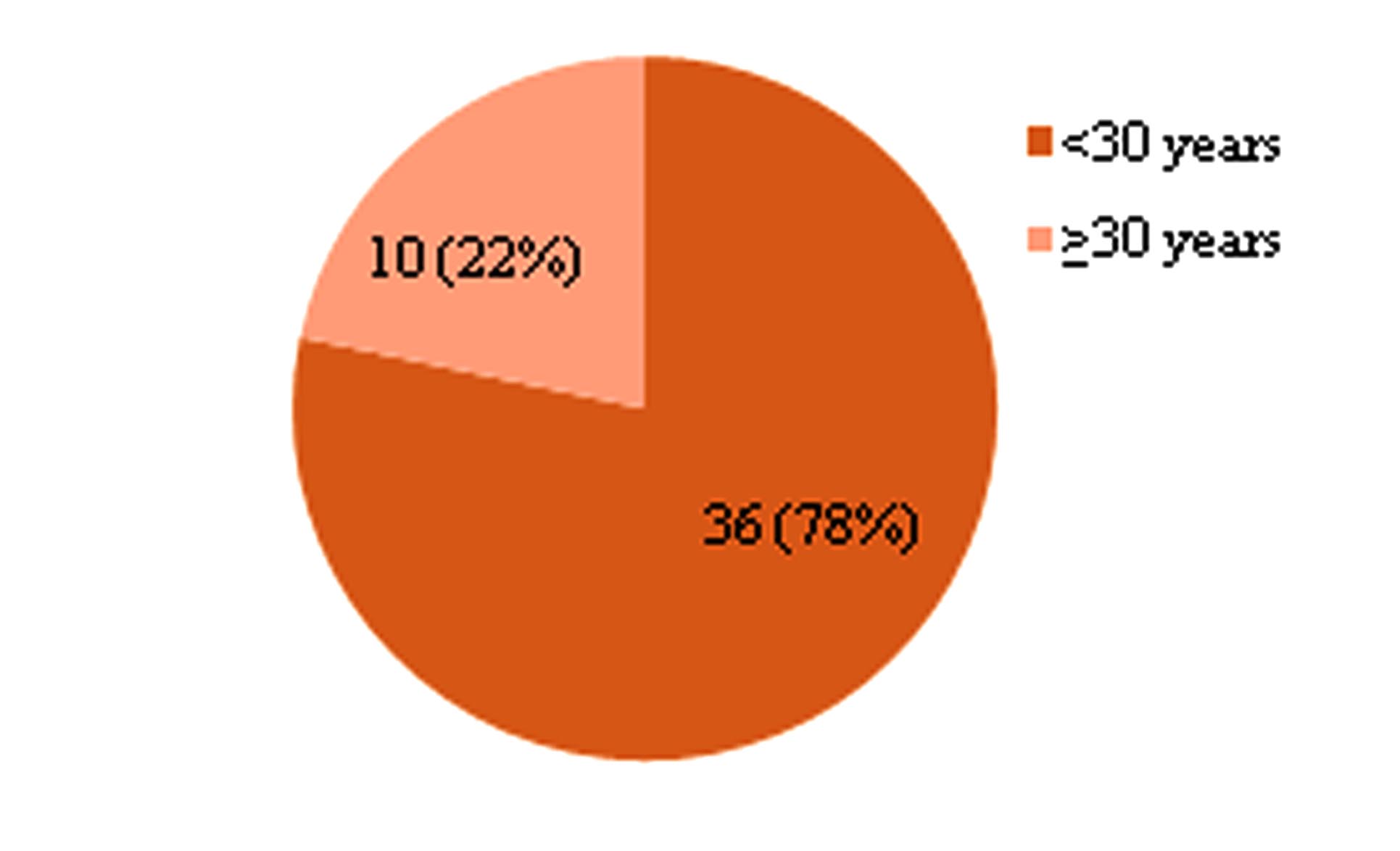
Forty six SLE cases were divided into two groups: first group less than 30 years and second group ≥30 years on the basis of their age of onset of disease. There were 36 (78%) cases in first group and 10 (22%) in second group in fig. 1
Out of 46 cases, the most frequently identified HLA-DR was DR2 (86.96%) followed by DR7 (43.48%), DR4 (17.39%), DR5 (17.39%), DR6 (13.04%), DR10 (13.04%), DR1 (4.35%) and DR3 (4.35%). HLA-DR8 and DR9 were not expressed by any cases. Out of 46 controls, the most frequently identified HLA-DR was DR2 (58.70%) followed by DR7 (41.30%), DR4 (21.72%), DR6 (19.57%), DR10 (19.57%), DR5 (10.87%), DR1 (8.70%), DR3 (8.70%), DR8 (8.70%) and DR9 (2.17%) fig 2.
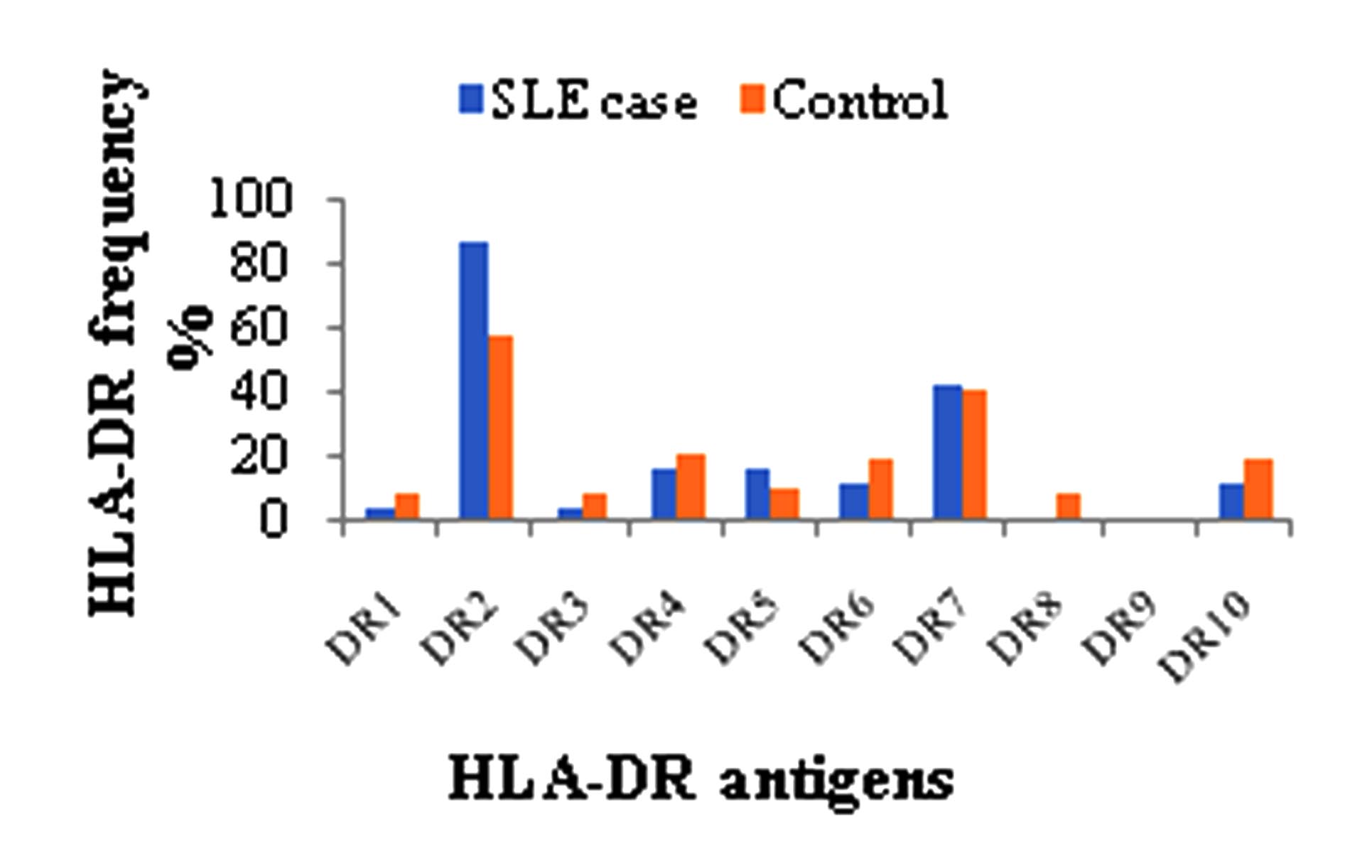
Positive association with SLE was observed for HLA-DR2 (86.96% Vs. 58.70%, p value = 0.0036, pc = 0.036, RR = 4.6914, 95% CI = 1.658 to 13.267) when compared with healthy controls. There were a weak increased of HLA-DR7 and HLA-DR5 in cases Vs. controls but it was not statistically significant at 5% signific-ance level. HLA-DR1, DR3, DR4, DR6, DR8, DR9 and DR10 were found to be slightly de-creased in cases as compared to controls. RR of these antigens was less than 1 but at 95% CI their association was not significant in table II
HLA-DR frequencies in cases that had disease onset less than 30 years of age were DR2 (86.11%), DR7 (44.44%), DR5 (19.44%), DR10 (13.89%), DR4 (16.67%), DR3 (2.78%), DR1 (5.56%) and DR6 (11.11%). In second group HLA-DR frequencies were DR2 (90%), DR7 (40%), DR4 (20%), DR6 (20%), DR3 (10%), DR5 (10%), and DR10 (10%). HLA-DR1 was not expressed by any cases in this group. In both groups no cases express DR8 and DR9 in table III
HLA-DR |
Cases |
Control |
p value |
pc |
RR |
95% CI |
DR1 |
2 (4.35) |
4 (8.70) |
0.4072 |
4.072 |
0.4773 |
0.083 to 2.744 |
DR2 |
40 (86.96) |
27 (58.70) |
0.0036* |
0.036* |
4.6914* |
1.658 to 13.267 |
DR3 |
2 (4.35) |
4 (8.70) |
0.4072 |
4.072 |
0.4773 |
0.083 to 2.744 |
DR4 |
8 (17.39) |
10 (21.72) |
0.5998 |
5.998 |
0.7579 |
0.269 to 2.134 |
DR5 |
8 (17.39) |
5 (10.87) |
0.3731 |
3.731 |
1.7263 |
0.519 to 5.739 |
DR6 |
6 (13.04) |
9 (19.57) |
0.3999 |
3.999 |
0.6167 |
0.200 to 1.900 |
DR7 |
20 (43.48) |
19 (41.30) |
0.8329 |
8.329 |
1.0931 |
0.478 to 2.499 |
DR8 |
0 (0) |
4 (8.70) |
0.1288 |
1.288 |
0.1016 |
0.005 to 1.942 |
DR9 |
0 (0) |
1 (2.17) |
0.4962 |
4.962 |
0.3262 |
0.012 to 8.217 |
DR10 |
6 (13.04) |
9 (19.57) |
0.3999 |
3.999 |
0.6167 |
0.200 to 1.900 |
|
≤30 years |
p value |
p corr |
RR |
≥30 years |
p value |
p corr |
RR |
DR1 |
2 (5.56) |
0.4460 |
4.460 |
1.5217 |
0 (0) |
0.44 |
4.4 |
0.657 |
DR2 |
31 (86.11) |
0.746 |
7.46 |
0.6889 |
9 (90) |
0.746 |
7.46 |
1.451 |
DR3 |
1 (2.78) |
0.909 |
9.09 |
0.2571 |
1 (10) |
0.909 |
9.09 |
3.888 |
DR4 |
6 (16.67) |
0.805 |
8.05 |
0.800 |
2 (20) |
0.805 |
8.05 |
1.250 |
DR5 |
7 (19.44) |
0.8216 |
8.216 |
2.1724 |
1 (10) |
0.821 |
8.21 |
0.460 |
DR6 |
4 (11.11) |
0.8355 |
8.355 |
0.500 |
2 (20) |
0.729 |
7.29 |
2.000 |
DR7 |
16 (44.44) |
0.8020 |
8.020 |
1.200 |
4 (40) |
0.802 |
8.02 |
0.833 |
DR8 |
0 (0) |
0 |
0 |
0 |
0 (0) |
0 |
0 |
0 |
DR9 |
0 (0) |
0 |
0 |
0 |
0 (0) |
0 |
0 |
0 |
DR10 |
5 (13.89) |
0.7467 |
7.467 |
1.451 |
1 (10) |
0.746 |
7.46 |
0.688 |
When comparison was made in these 2 groups, no positive association of HLA-DR with age of disease onset was found.
Discussion
Systemic lupus erythematosus (SLE) is a geneti-cally complex disease. The course of this chronic multisystem inflammatory disease may range from benign to fatal like organ failure (e.g., kid-ney) or malignancy. In spite of lots of study it is difficult to say the exact etiological factors caus-ing this disease. The importance of genetic fac-tors for development of disease has been con-firmed. But it is still controversial the contribu-tion of immunogenetic factors causing the dis-ease. A lot of work has been done to find out the degree and nature of association of human leukocyte antigen (HLA) and SLE. Genome wide association studies and haplotype study persuasively demonstrated the presence of SLE susceptibility factors in HLA-DRB1 and DQB1 alleles. Due to high degree of polymorphism within the genes of HLA, the distribution of HLA antigen is different according to race. Due to this variation, degree of association between SLE and specific genes of the major histocompatibility complex (MHC) also varies from one population group to another. This is the first description of immunogenetics of SLE in Bangladesh.A total 92 study population (46 cases & 46 con-trols) were enrolled in this study. Mean age of cases at study entry was 27.05 ± 8.17 years which is almost similar to the findings of other studies. Study by Sirikong et al., Hussain et al., Fouad et al. and Castano-Rodriguez et al. aver-age age was stated as 30.1 ± 10.5, 30.35 ± 1.687, 28 years and 34.7 ± 12.9 years in their studies, respectively.5,15-17 These findings indicate that SLE develops predominantly at 3rd and 4th decade of life. In this study, majority of the cases were female that was in agreement with other studies. Study among Pakistani population, Taiwan population, Northern Italian population, black South Africans and Malay people reported almost similar findings.16,18-21 All these studies indicate that SLE is more prevalent among fe-male. Higher prevalence of SLE in female could be due to direct effects of sex chromosomes or indirect effects of chromosomes such as those mediated by sex hormones.22 A recent study showed a 14 times higher incidence of SLE among Klinefelter’s, defined by 47, XXY karyo-types, than normal men. This risk (1: 900) is closer to the female risk of SLE (1: 1400 in Eu-ropean ancestry).23 The recent identification of IRAK1 gene supports direct effects of chromo-some X, possibly through gene dosage effects. In addition to IRAK1, a risk haplotype in the me-thyl-CpG-binding protein 2 gene (MECP2) has been associated with SLE and suggests a poten-tial role for DNA methylation in the pathogenesis of SLE. As with IRAK1, mapping of MECP2 on chromosome X raises the possibility of a gene dosage effect that may contribute to increased prevalence of SLE among women.22 Abnormal aestrogen metabolism has also been demonstrated in patients with SLE of both sexes. The con-centrations of androgens correlate inversely with disease activity. Excessive aestrogen causes pro-longed survival of autoimmune cells and increase Th2 cytokine production which in turn stimulates B cells to produce autoantibodies.12
In this study, the most frequent HLA-DR ob-served in cases was HLA-DR2. A positive asso-ciation of DR2 with SLE was observed when compared with controls. Similarly among Thais, Japanese, Malay, South Africans, Taiwanese and Kuwaiti populations with SLE, the frequency of expression of HLA-DR2 was more. In these stu-dies, positive association of HLA-DRB1*1502 (DR2) and DQB1*0501 among Thai population; HLA-DRB1*1501 (DR2), DRB5*0101 and DQB1*0602 among Japanese; HLA-DR2, DQB1*0501 and DQB1*0601 in Malay people; HLA-DR2 in black South Africans; HLA-DR2 among Kuwaiti population and Baltimore people; and HLA-DR2 among Chinese population have been reported.15, 17, 20, 21, 24-28 Haplotype analysis showed that haplotype containing DRB1*1501 and DRB1*0801 showed risk allele for SLE susceptibility and similar association was also showed in another study.7,29 Haplotype study in Saudis also showed association of HLA-DRB1*15 and DQB1*06 with SLE.30 Several other studies also showed association of HLA-DR2 with SLE in other populations such as study in Californiaand Latin American populations HLA-DR2 and DR3 showed the strongest association for susceptibility to SLE.5,31
But there are several studies that did not have similar findings with present study. Study among Pakistani population, western Indian, Egypt people showed different types of association other than HLA-DR2.16,32,33 Other studies among Norwegians, Spanish, Oklahoma, Toronto, UK, Southern Sweden and Netherland patients with SLE also showed positive association of DR3 with SLE.11,34-39 Study in Canada reported that among SLE patients with French origin HLA-DQ6 was positively associated and among non French Canadian SLE cases HLA-B8, DR3, Dw24 and DQ2 were associated.40 In all these studies, predominant HLA-DR among SLE patients was DR3 where as in the present study association of HLA-DR3 with SLE was not significant. A large series study among European SLE patients both HLA-DR2 and DR3 were found to be positively associated with SLE.41
A wide variation of negative association of HLA-DR with SLE was reported in many studies. A study among Toronto people reported a negative association of HLA-DR1, DR6 and DR7 with SLE.37 Negative association of HLA-DR7 with SLE in UK and HLA-DRB1*04, DRB1*07, DRB1*08 and DRB1*15 in Pakistani patients have also been reported.16,38 Some other studies also reported that HLA-DR9 and DR13 (split antigen of DR6) alleles may be protective alleles for Portuguese, Gypsies and Japanese people against SLE.24,42,43 A large series study of HLA class II alleles among European SLE patients showed negative association of some HLA class II antigens with SLE.41 Similarly negative association of HLA-DR5 with SLE among Latin American people has also been found.5 But in this study no HLA-DR was found to be negatively associated with SLE. Although, frequency of HLA-DR1, DR3, DR4, DR6, DR8, DR9 and DR10 decreased in cases compared to controls and their RR is less than 1 but at 95% CI their association was not significant (Table II). In future, if another study will do with large sample size these alleles may turn into negatively associated with SLE.
Some studies did not find any HLA-DR associa-tion with SLE. Association of HLA-DR with SLE among Jamaican, Iceland, Northern Italian, American Blacks, Chinese and Bulgarians SLE patients could not.19,44-48 These variations in the expression of HLA-DR among SLE patients re-ported in different studies and present study may be due to differences in ethnic, environmental exposure among study populations and con-founding factors in the extended MHC (xMHC) due to strong linkage disequilibrium.49 Although few studies showed an independent association of HLA class I with SLE, but the association was not consistent with the findings of other studies. Moreover, increase frequency of HLA-DR2 (58.70%) among control population compared to other DR in this study also indicate that these people may be at risk of development of SLE in future life if they exposed to appropriate envi-ronmental stimuli.
So, from the findings of above studies it has been revealed that some HLA alleles are protective and some are associated with increased risk for development of SLE. Several studies have been carried out to understand the mechanisms underlying MHC association with SLE, but yet not clearly understood. But it has been hypothesized that physiochemical differences in the nature of critical amino acid side chains that shape the peptide-binding groove in the DRβ chain might be related to the risk or protection conferred by HLA-DRB1 alleles associated with SLE.5 In this study, it was already reported that, the cases were divided into 2 groups according to their age of onset of disease shown in fig. 1. When frequency of HLA-DR was compared among 2 groups, both groups have almost same frequency of HLA-DR irrespective of their age of onset of disease. Although frequency of HLA-DR2 was increased in second group but it was not statistically significant. Similarly, study in Malay people any HLA-DR association with age of onset of disease could not confirmed.21 But another study reported increased frequency of DR3 bearing haplotype in patients with disease onset before 30 years of age compared to those above 30 years of age.50 This could be explained by the difference in race/ ethnic of study popula-tions and small sample size. To get a better pic-ture regarding association of HLA with age of onset of disease large sample size from multicen-ter analysis is needed.
Thus, the ethnic difference among study popula-tions, sample size of different study and hetero-genecity of the HLA could explain the contradic-tory results found in different studies mentioned above.
Therefore, the results of this study further con-firmed that HLA-DR particularly DR2 has role in development of SLE, may be by influencing production of specific autoantibodies. But it also has been observed that onset of SLE is more likely the consequence of cooperation of many other genes including HLA-DR, interacting with appropriate external stimuli. Because of small sample size, the study population in this study may not represent the entire SLE cases in Bangladesh. This study only included certain serological types of HLA. As HLA-DR is in linkage disequilibrium with other HLA, study of other loci of HLA at allele level thus recommend for proper identification of HLA association with SLE.
Acknowledgement:
Authors are thankful to Dept. of Rheumatology, BSMMU and Dept. of Micro-biology and Immunology for their support to conduct the study. Authors are also thankfulto the lupus patients and all the medical persons who were control population, for their coopera-tion and support that made this study possible.
References
- Bongu A, Chang E and Goldman RR. Can morbidity and mortality of SLE be improved? Best practice and research; clinical rheumatology 2002; 16 (2): 313-32.
- Lau CS, Yin G, and Mok MY. Ethnic and geographi-cal differences in systemic lupus erythematosus: an overview. Lupus 2006; 15(11): 713-14.
- Deapen D, Escalante A, Weinrib L et al., A revised estimate of twin concordance in SLE. Arthritis and Rheumatism 1992; 35: 311-18.
- Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH et al. Familial aggregation of systemic lupus ery-thematosus, rheumatoid arthritis and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis and Rheumatism 2005; 52(4): 1138-47.
- Castano- Rodriguez N, Diazo- Gallo LM, Pineda- Tamayo R et al. Meta- analysis of HLA-DRB1 and HLA-DQB1 polymorphisim in Latin American pa-tients with systemic lupus erythematosus, Autoim-munity Reviews 2008; 7: 322-30.
- Forabosco P, Gorman JD, Cleveland C et al. Meta-analysis of genome-wide linkage studies of systemic lupus erythematosus, Genetics and immunity 2006; 7(7): 609-14.
- Graham RR, Ortmann WA, Rodine P et al. Specific combinations of HLA-DR and DR3 class 11 haplo-types contribute graded risk for disease susceptibility and autoantibodies in human SLE, European journal of human genetics 2007; 15: 823-30.
- Grumet FC, Coukell A, Bodmer JG, Bodmer WF, McDevitt HO. Histocompatibility (HL-A) antigens associated with systemic lupus erythematosus. A possible genetic predisposition to disease. The New England Journal of Medicine 1971; 285: 193-96.
- Pisetsky DS. Systemc lupus erythematosus. A. Epi-demiology, pathology and pathogenesis. In: Klippel JH, ed. Prime on the rheumatic diseases, 11th ed. Georgia, USA: Arthritis Foundation 1997: 246-51.
- D'Alfonso S, Rampi M, Bocchio D et al. Systemic lupus erythematosus candidate genes in the Italian population: evidence for a significant association with interleukin-10. Arthritis Rheumatism 2000; 43(1): 120-8.
- Rood MJ, van Krugten MV, Zanelli E et al. TNF-308A and HLA-DR3 alleles contribute independently to susceptibility to systemic lupus erythematosus. Arthritis Rheumatism 2000; 43(1): 129-34.
- Mok CC, Lau CS. Pathogenesis of systemic lupus erythematosus. Journal of Clinical Pathology 2003; 56: 481-90.
- Gill JM, Quisel AM, Rocca PV, Walters DT. Diagno-sis of Systemic Lupus Erythematosus. American Family Physician 2003; 68 (11): 2179-86.
- uenaga E and Nakamura H. Evaluation of three me-thods for effective extraction of DNA from human hair. Journal of Chromatography B 2005; 820: 137-41.
- Sirikong M, Tsuchiya N, Chandanayingyong D et al., Association of HLA-DRB1*1502-DQB1*0501 hap-lotype with susceptibility to systemic lupus erythema-tosus in Thais. Tissue Antigen 2002; 59: 113-17.
- Hussain N, Jaffery G, Sabri AN, Hasnain S. HLA Association in SLE patients from Lahore-Pakistan. Bosnian Journal of Basic Medical Sciences 2011; 11 (1): 20-26.
- Fouad F, Johny K, Kaaba S, Alkarmi TO, Sharma P & Al-Hrbi S. MHC in systemic lupus erythematosus: A study on a Kuwaiti population. European Journal ofImmunogenetics 1994; 21: 11-14.
- Ling- Ying LU, Ding WZ, Deulofeut R et al., Mole-cular analysis of major histocompatibility complex al-lelic associations with systemic lupus erythematosus in Taiwan. Arthritis and Rheumatism 1997; 40(6): 1138-45.
- Savi M, Ferraccioli GF, Neri TM et al., HLA-DR an-tigens and anticardiolipin antibodies in Northern Italian systemic lupus erythematosus patients. Arthritis and Rheumatism1988; 31(12): 1568-70.
- Rudwaleit M, Tikly M, Gibson K, Pile K, Wordsworth P. HLA class II antigens associated with systemic lupus erythematosus in black South Africans. Annals ofRheumatic Diseases 1995; 54: 678-80.
- Azizah MR, Ainol SS, Kong NC, Normaznah Y, Ra-him MN. HLA antigens in Malay patients with sys-temic lupus erythematosus: association with clinical and autoantibody expression. The Korean Journal of Internal Medicine 2001; 16(2): 123-31.
- Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus ery-thematosus. Genes and Immunity 2009; 10: 373-79.
- Scofield RH, Bruner GR, Namjou B et al., klinefel-ter’s syndrome (47,xxy) in male systemic lupus ery-thematosus patients. Arthritis & Rheumatism 2008; 58(8): 2511-17.
- Shimane K, Kochi Y, Suzuki A et al., An association analysis of HLA-DRB1 with systemic lupus erythe-matosus and rheumatoid arthritis in a Japanese popu-lation: effects of *09:01 allele on disease phenotypes. Rheumatology 2013; 52(7): 1172-82.
- Furukawa H, Kawasaki A, Oka S et al., Human leu-kocyte antigens and systemic lupus erythematosus: A protective role for the HLA-DR6 alleles DRB1*13:02 and *14:03. PlosOne 2014; 9(2): e87792.
- Hochberg MC, Boyd RE, Ahearn JM et al., Systemic lupus erythematosus: A review of clinic-laboratory features and immunogenetic markers in 150 patients with emphasis on demographic subsets. Medicine 1985; 64(5): 285-95.
- Mohd-Yusuf Y, Phipps ME, Chow SK and Yeap SS. HLA-A*11 and novel associations in Malays and Chinese with systemic lupus erythematosus. Immu-nology 2011; 139: 68-72.
- Graham RR, Ortmann WA, Langefeld CD et al., Vi-sualizing human leukocyte antigen class II risk haplo-type in human systemic lupus erythematosus. Am. J. Hum. Genet 2002; 71: 543-53.
- Al-motwee S, Jawdat D, Jehani GS et al., Association of HLA-DRB1*15 and HLA-DQB1*06 with SLE in Saudis. Annals of Saudi Medicine 2013; 33(3): 229-34.
- Fronek Z, Timmerman LA, Alper CA et al., Major histocompatibility complex genes and susceptibility to systemic lupus erythematosus. Arthritis and Rheu-matism 1990; 33(10): 1542-53.
- Shankarkumar U, Ghosh K, Badakere SS, Mohanty D. HLA-DRB1*03 and DQB1*0302 associations in a subset of patients severely affected with systemic lu-pus erythematosus from western India. Annals of the RheumaticDiseases 2003; 62: 92-3.
- Farouk HM, Gabal MMA, Ali AM, Al-Hefny AAM, Fahmy HM, Zarouk WA. Contribution of Human Leucocyte Antigen HLA-DR in Systemic Lupus Ery-thematosus: Effect on Disease Susceptibility and Outcome. Research Journal of Medicine andMedical Sciences 2011; 6(1): 43-50.
- Skarasvag S, Hansen KE, Moen T and Eggen BM. Distribution of HLA class II alleles in autoantibody subsets among Norwegain patients with systemic lu-pus erythematosus. Scand. J. Immunol 1995; 42: 564-71.
- Martin-Villa JM, Martinez-Laso J, Moreno-Pelayo MA et al., Differential contribution of HLA-DR, DQ and TAP2 alleles to systemic lupus erythematosus susceptibility in Spanish patients: role of TAP2*01 alleles in Ro autoantibody production. Annals of the RheumaticDiseases 1998; 57: 214-19.
- Smolen JS, Klippel JH, Penner E et al., HLA-DR an-tigens in systemic lupus erythematosus: association with specificity of autoantibody response to nuclear antigens. Annals of the RheumaticDiseases 1987; 46: 457-62.
- Gladman DD, Urowitz MB and Darlington GA. Dis-ease expression and class II HLA antigens in systemic lupus erythematosus. Lupus 1999; 8: 466-70.
- McHugh NJ, Owen P, Cox B et al., MHC class II, tumor necrosis factor α, and lymphotoxin α gene hap-lotype associations with serological subsets of sys-temic lupus erythematosus. Annals of the Rheuma-ticDiseases 2006; 65: 488-94.
- Tjernstrom F, Hellmer G, Nived O et al., Synergetic effect between interleukin-1 receptor antagonist allele (IL1RN*2) and MHC class II (DR17, DQ2) in deter-mining susceptibility to systemic lupus erythematosus. Lupus 1999; 8: 103-08.
- Goldstein R and Sengar DPS, Comparative studies of the major histocompatibility complex in French Ca-nadian and Non French Canadian Caucasians with systemic lupus erythematosus. Arthritis and Rheu-matism 1993; 36(8): 1121-27.
- Galeazzi M, Sebastiani GD, Morozzi G et al., HLA class II DNA typing in a large series of European pa-tients with systemic lupus erythematosus. Medicine 2002; 81: 169-78.
- Vasconcelos C,Carvalho C, Leal B et al.,HLA in Por-tuguese Systemic Lupus Erythematosus Patients and Their Relation to Clinical Features. Annals of the New YorkAcademy of Sciences 2009; 1173: 575-80.
- Ramal LM, Lopez-Nevot MA, Sabio JM et al., Sys-temic lupus erythematosus in southern Spain: a com-parative clinical and genetic study between Caucasian and Gypsy patients. Lupus 2004; 13: 934-40.
- Christian N, Smikle NF, DeCeulaer K, Daniels L, Walravens MJ and Barton EN. Antinuclear antibodies and HLA class II alleles in Jamaican patients with systemic lupus erythematosus. West Indian Med J 2007; 56: 130-33.
- Steinsson K, Jonsdottir S, Arason GJ et al., A study of the association of HLA DR, DQ and complement C4 alleles with systemic lupus erythematosus in Icel-and. Annals ofRheumatic disease 1998; 57: 503-5.
- Reveille JD, Schrohenloher RE, Acton RT and Barger BO. DNA analysis of HLA-DR and DQ genes in American Blacks with systemic lupus erythematosus. Arthritis andRheumatism 1989; 32(10): 1243-51.
- Savage DA, Ng SC, Howe HS, et al., HLA and TAP associations in Chinese SLE. Tissue Antigens 1995; 46: 213-16.
- Marintchev LM, Naumova EJ, Rashkov RK, Arnett EC, Reveille JD. HLA class II alleles and autoanti-bodies in Bulgarians with SLE. Tissue Antigen 1995; 46: 422-25.
- Smerdel-Ramoya A, Finholt C, Lilleby V et al., Sys-temic lupus erythematosus and the extended major histocompatibility complex—evidence for several predisposing loci. Rheumatology 2005; 44: 1368-73.
- Davies EJ, Hillarby MC, Cooper RG et al., HLA-DQ, DR and complement C4 variants in SLE. Br J Rheu-matol 1993; 32: 870-75.
- Soc Neonatal 2012; 19(4): 229-236
24. Hashimoto H, Nishimura Y, Dong RP et al., HIA An-tigens in Japanese Patients with Systemic Lupus Ery-thematosus. Scand J Rheumatol 1994; 23: 191-96.