Shamsun Nahar
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Umme Laila Urmi
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Tamanna Ali
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Adity Rumnaz
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Tanjum Ara Haque
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Bayasrin Ara
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Mohammed Shah Alam
Department of Microbiology, Gono Bishwabidyalay, Savar, Dhaka-1344, Bangladesh.
Abu Syed Md. Mosaddek
Department of Pharmacology, Uttara Adhunik Medical College, Uttara, Dhaka, Bangladesh.
Salequl Islam
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Keywords: Antibiotic resistance, ESBL genes, blaTEM, blaOXA, blaSHV.
DOI: 10.3329/bmrcb.v47i2.57775
Abstract
Background: In Bangladesh, the poultry industry contributes a significant role in the food sector. A vast amount of antibiotic is used as prophylaxis and growth promotion factors in farms. These unconcerned uses of antibiotics ultimately generate resistant bacteria affecting substantial adverse consequences on human health. Extended-spectrum b-lactamase (ESBL) genes are responsible for inactivation of antibiotics containing b-lactam ring, namely, penicillin, cephalosporins, monobactams, and carbapenems.
Objectives: This study was designed to analyse the distribution of three ESBL genes and associated antimicrobial susceptibility profile of poultry gut bacteria.
Methods: This study was designed to analsze the distribution of three ESBL genes and associated antimicrobial susceptibility profile of poultry gut bacteria. Poultry feces were collected and cultured on cysteine lactose electrolyte deficient (CLED) agar and Salmonella-Shigella (SS) agar to differentiate various isolates based on colony characteristics. Identification of the isolates was made by convention biochemical tests, analytical profile index (API-20E), and 16S rRNA sequence analysis. Antibiotic susceptibility test was done by disc diffusion method using 17 antibiotics from seven groups. Subsequently, polymerase chain reaction (PCR) was employed with a specific primer to identify respective ESBL genes (blaTEM, blaSHV, blaOXA). All data were analysed by SPSS.
Results: A total of 113 isolates were identified from 85 poultry feces tested. Most of the bacteria belonged to Enterobacteriaceae family, notably Proteus spp., E. coli, Klebsiella spp., Salmonella spp., and Enterobacter spp. Different bacteria were detected, namely, Kurthia populi, Cronobactersp, and Eikenella corrodens. Most of the poultry isolates were resistant against more than one group of antibiotics. ESBL gene, blaTEM gene was identified most frequently (53.9%), followed by blaOXA (52.2%), and blaSHV (23%). Higher phenotypic resistance was observed in isolates carrying ESBL genes.
Conclusion: This study revealed a very high frequency of three ESBL genes with their phenotypic resistance- capacities in Bangladeshi poultry gut microbiota. Excess uses of antibiotics in local poultry farms may result in the emergence of antibiotic resistance that is imposing public health threatening in Bangladesh.
Keywords: Antibiotic resistance, ESBL genes, blaTEM, blaOXA, blaSHV.
Introduction
Antimicrobial resistance (AMR) has become a rapidly growing public health concern worldwide. Infections from resistant bacteria are now too common, and some pathogens have even become resistant to multiple types of antibiotics.1,2 Non-judicial use of antibiotics is considered to be the most significant reason for the emergence, selection, and spreading of antibiotic-resistant bacteria in both animals and humans.3,4 Antibiotics are used extensively in the poultry industry for therapeutic and prophylactically, including growth promoters in poultry feeds.5 Aggressive use of antibiotics plays a crucial role in the emergence of antibiotic resistance among gram- negative bacteria worldwide, and limiting treatment options considerably.6 Recent studies have recognized that inappropriate use of antibiotics in animal husbandry leads to the increase of multidrug- resistant bacteria and also considered as a potential reservoir of resistant bacteria.7,8,9 Antibiotics can lead to the emergence and dissemination of different resistant bacteria, which can be passed on to people via food or direct contact with infected animals.10,11
Beta-lactams are the most widely used antibiotics, including natural and synthetic penicillins and their derivatives such as cephalosporins, cephamycins, monobactams, and carbapenems.12 Resistance to beta-lactam antibiotics is of special concern because of their critical importance for humans and veterinary medicine.13 Food-producing animals, especially poultry, have been suggested as a potential source for transmission of extended-spectrum beta-lactamase (ESBL)-producing bacteria to humans, either by direct contact or consumption of contaminated meat products, leading to the colonization of the intestinal tract and eventually to severe infections.14 Human infection due to ESBL producing bacteria is associated with increased mortality, morbidity, high cost of hospitalization, and delay appropriate therapy.15
ESBLs are most commonly found in Entero- bacteriaceae. Escherichia coli and Salmonella spp. are often common ESBL-producers isolated in poultry and its environment.16 These bacteria are resistant to penicillins, cephalosporins, and aztreonam mainly due to the production of CTX-M, TEM, and SHV b-lactamases, which are encoded by blaCTX-M, blaSHV, and blaTEM genes, respectively.17,18
There is limited information about the ESBL gene carriage in Bangladeshi animal husbandry, particularly in poultry-gut bacteria. Poultry feces are a very prominent source for analyzing antibiotic-resistant bacteria and the ESBL genes. The main objective of the present study was to detect the type of bacteria in poultry feces with the extent of their antibiotic resistance and the presence of three ESBL genes, namely, blaSHV, blaTEM and blaOXA. Commensal bacteria represent a reservoir for these antibiotic resistance genes, which can be disseminated into different other recipient bacteria progressively.19 This study unveils the local distribution of the three ESBL genes, blaSHV, blaTEM, and blaOXA among isolates from poultry feces in Bangladesh. Thus, the research contributes to generating some evidence-based information about the reservoir of antimicrobial resistance in a food-animal industry.
Materials and Methods
Study Design and Specimen Collection: A cross- sectional study was conducted between July 2019 and December 2019 to examine ESBL gene prevalence in bacterial isolates from chicken droppings. Poultry farms (PFs) were conveniently selected by the trained team of microbiologists, veterinary doctors, clinicians, public health professionals, statisticians as well as postgraduate students for the chicken-feces collection followed by microbiology analyses. We selected 17 PFs from areas of Savar, Hemayetpur, Manikganj, Gazipur, Tangail, and Mymensingh, where the major poultry industry is located in Bangladesh. Geographic information mapping software, ArcGIS version 10 for Windows, was used to draw a sampling spot-location map (Figure 1). A structured questionnaire was approached to farm owners to enquire types of poultry chicken, their recent disease history, and records of antibiotics applied for treatment and/or prophylaxis. The query had also sought the farm-owners about their levels of education and attained training on animal husbandry.
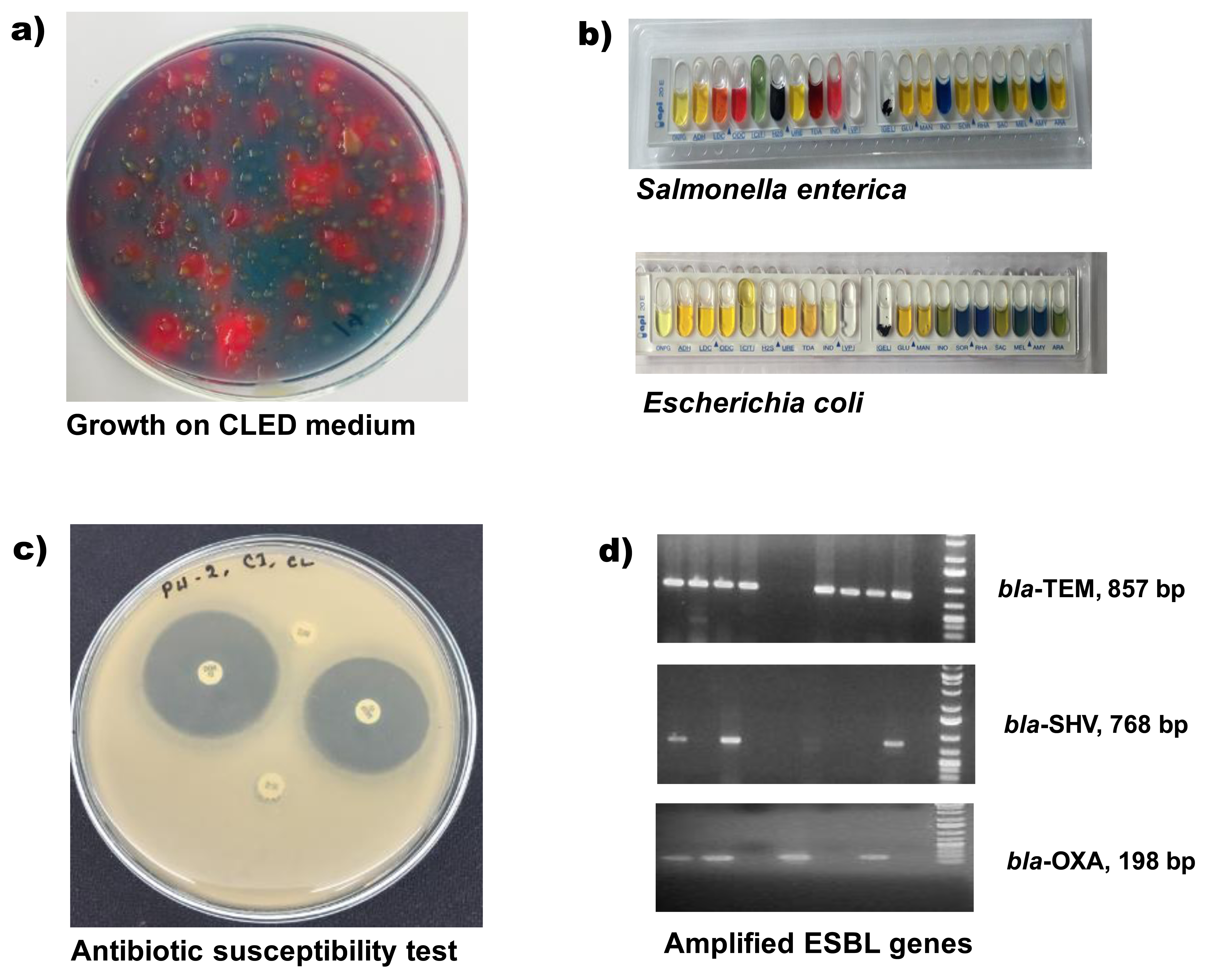
Bacterial isolation and Iidentification: Chicken faecal samples were directly collected in specific specimen collection tube following all safety precautions and aseptic techniques. Samples were stored immediately in insulated ice-boxes and transported to the One Health Laboratory at the Department of Microbiology, Jahangirnagar University, Bangladesh. All associated microbiological and molecular biology analyses were carried out there. Long distant feces samples were dipped into Cary Blair transport medium (Oxoid, UK) before shifting into the laboratory. For bacterial isolation, approximate one gram of chicken- feces was mixed in four mL of phosphate-buffered saline (PBS), and one loopful diluted sample was streaked on a differential culture medium, cysteine-, lactose-, and electrolyte-deficient (CLED) agar (Lyophilchem, Italy) for growth of Gram-negative enteric bacilli. For the detection of Salmonella and Shigella, the diluted chick-droppings were enriched in Rappaport Vassiliadis Soya Broth (RVS Broth, Oxoid, UK) and streaked separately on Salmonella-Shigella (SS) agar (Oxoid, UK) media. After overnight incubation at 37°C, each type of bacteria was differentiated initially on their colony characteristics (figure 2a). A distinct single colony was picked-up and cultured again on tryptone soya agar (Lyophilchem, Italy) for preparing pure- culture repository and for further analyses. The purified bacterial colonies were identified by conventional biochemical procedures followed by a rapid biochemical-test kit (API 20E, BioMe´rieux, Durham, NC) consisting of a set of the chromogenic panel, carbohydrate batteries, and enzymatic substrates (Figure 2).20
Antimicrobial susceptibility testing: Antimicrobial susceptibility testing was performed by the disc diffusion method (Kirby-Bauer disc diffusion method) on Mueller-Hinton agar (MHA) plates and the zone diameter for individual antimicrobial agents interpreted according to Clinical Laboratory Standards Institute recommendations (CLSI 2016) and then translated into sensitive, moderate or resistant categories (figure 2c).21 Bacillus cereus ATCC 14579 was used as the quality control strain. Seventeen different commercially available antibiotic discs (Oxoid, Basingstoke, United Kingdom) belonging to seven individual groups (Beta-lactam, Carbapenem, Sulphamethaxazol Trimethoprim, Nitrofurantoin, Fluoroquinolones, Aminoglycoside, and Macrolides) of antibiotics were used for the test. The utilized antimicrobials included Amoxycillin+Clavulinic acid (20+10ìg), Cephalexin (30ìg), Cefuroxime sodium (30ìg), Cefixime (5ìg), Ceftriaxone (30ìg), Cefepime (30ìg), Imipenem (10ìg), Sulphamethaxazol Trimethoprim (25ìg), Nitrofurantoin (100ìg), Ofloxacin (5ìg), Lomefloxacin (10ìg), Nalidixic Acid (30ìg), Ciprofloxacin (5ìg), Gentamycin (10ìg), Amikacin (30ìg), Netlimycin (30ìg) and Azithromycin (15ìg).
Detection of ESBL specific genes: The conventional polymerase chain reaction (PCR) method was applied for screening of all isolates for the presence of blaTEM, blaOXA, and blaSHV genes. The sequences of primers used in this study and specific for blaTEM, blaOXA, and blaSHVwere listed in Table I. For PCR, freshly cultured isolates bacteria were used to prepare template deoxyribonucleic acid (DNA) by the boiling method.22 For each PCR reaction, prepared bacterial DNA 2.0 µL was added to a 12 µL 2X PCR pre-mixture (GeneON, Germany) and five pmol of each primer (1 µL), and the remaining deionized water to make a final volume of 24 µL. Reactions underwent an initial denaturation at 95°C for 10 min followed by 32 cycles of amplification (Applied Biosystems 2720 Thermal Cycler, Singapore), consisting of denaturation 30s at 94 °C, annealing 30s at 52 °C, extension 1 min at 72°C, and a final 7 min extension at 72°C. Amplicons (857bp, 198bp, and 768bp for blaTEM, blaOXA, and blaSHV, respectively) were visualized under UV light after electrophoresis through 1.2% agarose gel at 100 volts for 30 minutes, followed by staining with ethidium bromide. The standard molecular weight marker (GeneRuler, ThermoFisher Scientific, MA) was run parallel to measure specific amplicon sizes (figure 2d).
Target gene |
primer |
sequence (5’-3’) |
Amplicon size |
References |
|---|---|---|---|---|
blaTEM |
Forward |
GAGTATTCAACATTTTCGT |
857 bp |
(Van et al., 2008) |
|
Reverse |
ACC AATGCTTAATCAGTGA |
|
|
blaOXA |
Forward |
GCAGCGCCAGTGCATCAAC |
198 bp |
(Van et al., 2008) |
|
Reverse |
CCGCATCAAATGCCATAAGTG |
|
|
blaSHV |
Forward |
TCGCCTGTGTATTATCTCCC |
768 bp |
(Van et al., 2008) |
|
Reverse |
CGCAGATAAATAACCACAATG |
|
|
Statistical Analysis: Data were verified, entered, and subsequently analyzed using IBM SPSS statistics data editor. Missing data were omitted from the bivariate analysis.
This study was approved by the National Research Ethics Committee (NREC) of the Bangladesh Medical Research Council (BMRC) [BMRC/Grants/2018-2019/ 99, dated 31.10.2018]. Verbal consents were obtained from poultry farm owners and homeowners for collecting the respective chicken droppings.
Results
Study Farms and Samples: Small- and medium-scale poultry farms (PFs) have been expanded extensively in commercial and traditional levels in Bangladesh. A total of 85 feces housed in 17 poultry farms in six districts of Bangladesh were screened for distinct types of enteric bacteria (figure 1).
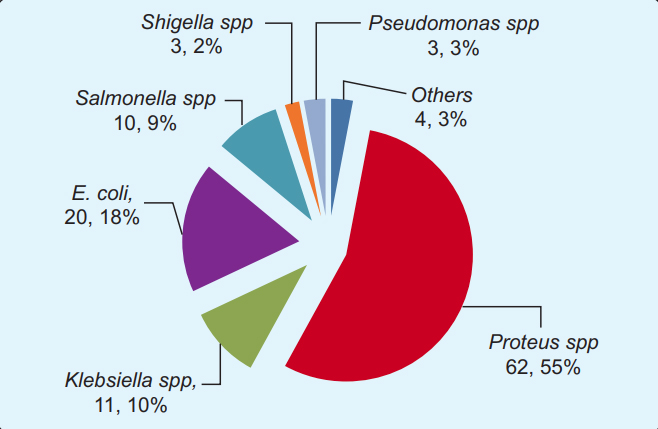
Isolation and Identification of Chicken Feces Bacteria: A total of 113 isolates were yielded from the 85 poultry feces examined. As a whole, all the poultry feces yielded at least one or more types of bacteria. Very few culture plates appeared no growth, where we repeated the procedure the next day from preserved samples to validate prior results. Any discordant of the two culture- results were excluded from the analysis. The 113 poultry faeces isolates were classified as, 20 E. coli, 62 Proteus spp., 11 Klebsiella spp., 10 Salmonella spp., three Shigella spp., and three Pseudomonas spp. Four different bacteria, namely Enterobacter, Kurthia populi, Cronobacter, and Eikenella corrodens had also been identified in (Figure 3).
Antibiotic Susceptibility Profiles of Isolates: The isolates were subjected to antimicrobial susceptibility tests against 17 different antimicrobial agents of seven groups in order to evaluate their resistance patterns. The phenotypic susceptibilities of the isolates against the different tested antimicrobial agents have been summarised (table II).
Group |
Antibiotics |
Susceptibility among tested isolates, n (%) |
Pseudomonas Pseudomonas (n=3) |
Others (n=4) |
||||
|---|---|---|---|---|---|---|---|---|
Proteus spp. (n=62) |
Klebsiella spp. (n=11) |
Salmonella spp.(n=10) |
Shigella spp. (n=3) |
E. coli (n=20) |
||||
b-lactam |
Amoxycillin-Clavulanate |
2 (3.2) |
1 (9.1) |
1(10) |
1(33.3) |
0 (0) |
0 (0) |
0 (0) |
Cephalexin |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|
Cefuroxime |
6 (9.6) |
1 (9.1) |
1 (10) |
0 (0) |
1(5) |
0 (0) |
0 (0) |
|
Cefixime |
4 (6.4) |
0 (0) |
0 (0) |
0 (0) |
2 (10) |
0 (0) |
0 (0) |
|
Ceftriaxone |
32 (51.6) |
4 (36.3) |
2 (20) |
1 (33.3) |
15 (75) |
0 (0) |
1 (25) |
|
Cefepime |
6 (9.6) |
1 (9.1) |
1 (10) |
1 (33.3) |
2 (10) |
0 (0) |
0 (0) |
|
Imipenem |
11 (17.74) |
1 (9.09) |
2 (20) |
0 (0) |
6 (30) |
1(33.3) |
1(25) |
|
Co-trimoxazole |
TMP/SMX a |
1 (1.6) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
Nitrofuran |
Nitrofurantoin |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
Quinolone/ Fluoroquinolone |
Ciprofloxacin |
5 (8.7) |
0 (0) |
0 (0) |
0 (0) |
6 (30) |
0 (0) |
0 (0) |
Nalidixic Acid |
1(1.6) |
0 (0) |
0 (0) |
0 (0) |
1 (5) |
0 (0) |
0 (0) |
|
Ofloxacin |
2 (3.2) |
0 (0) |
0 (0) |
1(33.3) |
3 (15) |
1(33.3) |
0 (0) |
|
Lomefloxacin |
0 (0) |
0 (0) |
0(0) |
0 (0) |
1(5) |
0 (0) |
0 (0) |
|
Aminoglycoside |
Gentamycin |
26 (41.9) |
2 (18.2) |
1(10) |
3 (100) |
15 (75) |
0 (0) |
0 (0) |
Amikacin |
34 (54.8) |
4 (36.4) |
4 (40) |
3 (100) |
10 (50) |
1(33.3) |
1(25) |
|
Netilmicin |
22 (35.4) |
0 (0) |
1(10) |
1(33.3) |
5 (25) |
1 (33.3) |
1(25) |
|
Macrolides |
Azithromycin |
12 (19.4) |
1(9.1) |
1(10) |
1(33.3) |
3 (15) |
1(33.3) |
1(25) |
The majority of the isolates exerted resistance to more than one group of antibiotics. The result of disc diffusion susceptibility testing revealed that all of the Proteus spp. were resistant to Cephalexin, Nitrofurantoin, and Lomefloxacin. The susceptibility rate of isolated Proteus spp. to other antibiotics was: Ceftriaxone 51.6%; Amikacin 54.8%; Gentamycin 41.9%; Netlimycin 35.4%. Less than 20% of Proteus spp, isolates were sensitive to all other tested antibiotics.
Besides resistance to all antibiotics of fluoroquinolone group, Klebsiella spp also resistant to Cephalexin, Cefuroxime sodium, Sulphamethaxazol-Trimethoprim, Nitrofurantoin, and Netlimycin. About 36% of Klebsiella spp. were sensitive to Ceftriaxone and Amikacin. Besides against Gentamicin (18.2%), less than 10% sensitivity was shown by all of the Klebsiella spp. against tested antibiotics.
All of the isolated Salmonella spp. was resistant to Cephalexin, Cefixime, Sulphamethaxazol Trimethoprim, Nitrofurantoin, and all antibiotics of fluoroquinolone group. Around 40% of Salmonella spp. were sensitive to Amikacin, 20% sensitive to Ceftriaxone, and Imipenem and 10% sensitive against the rest of the tested antibiotics. In the case of isolated Shigella spp., sensitivity was 100% against Gentamycin and Amikacin, whereas 33.3% isolates were sensitive against Amoxycillin+Clavulinic acid, Ceftriaxone, Cefepime, Ofloxacin, Netlimycin, and Azithromycin.
Furthermore, the results of the antimicrobial susceptibility test revealed that E. coli showed 100% resistance to Amoxicillin+Clavulanic acid, Cephalexin, Sulphamethaxazol-Trimethoprim, and Nitrofurantoin. However, 75% of the isolates showed susceptibility against Ceftriaxone and Gentamycin, 50% against Amikacin, and 30% against ciprofloxacin and Imipenem. Only 5–25% E. coli showed susceptibility to all Cefuroxime, Cefixime, Cefepime, Ofloxacin, Lomefloxacin, Nalidixic Acid, Netlimycin, Azithromycin. Except for Ofloxacin, all antibiotics of fluoroquinolone and beta-lactam groups showed resistance to all isolated Pseudomonas spp. Over 33.0% of isolates remained sensitive to all other tested antibiotics. Over all 25% of Pseudomonas spp. isolates were sensitive against Ceftriaxone, Imipenem, Amikacin, Netlimycin, and Azithromycin.
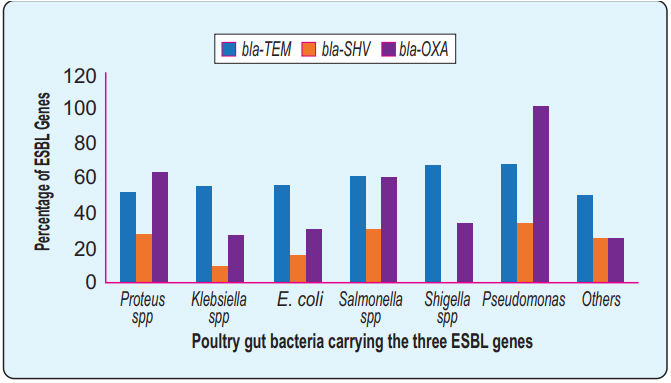
Distributions of the ESBL genes: The PCR data revealed that the blaTEM gene was the most frequent (53.9%) followed by the blaOXA (52.2%) and finally the blaSHV (23%) respectively. The overall frequency of ESBL genes among different bacteria (Figure 4). None of the Shigella spp. contained a blaSHV gene where the blaOXA gene detected in all Pseudomonas spp. 62.9% Proteus spp contained blaOXA gene besides 54.5% Klebsiella spp, and 55% E. coli contain blaTEM gene.
Association of blaTEM, blaSHV, and blaOXA: Three different ESBL gene variants were detected. blaTEM was found to be more prevalent, followed by blaOXA. Association of the three ESBL genes to their phenotypic resistance to b-lactam group antibiotics (table III).
ESBL |
|
Genes% of phenotypic resistance to b-lactam antibiotics |
||||||
|---|---|---|---|---|---|---|---|---|
Amoxiclav (30µg) |
Cephalexin (30µg) |
Cefuroxime (30µg) |
Cefixime (5µg) |
Ceftriaxone (30µg) |
Cefepime (30µg) |
Imipenem (10µg) |
||
bla-TEM |
Presence (n=61) |
95 |
100 |
91 |
95 |
42.62 |
90 |
82 |
|
Absence (n=52) |
96 |
100 |
92 |
94 |
61.5 |
90 |
79 |
bla-SHV |
Presence (n=26) |
96 |
96 |
84.6 |
100 |
50 |
88 |
65 |
|
Absence (n=87) |
95 |
100 |
94 |
93 |
51.7 |
90.8 |
80 |
bla-OXA |
Presence (n=59) |
96.6 |
100 |
89.8 |
96.6 |
61 |
89.8 |
86 |
|
Absence (n=54) |
94 |
100 |
94 |
92.5 |
40.7 |
90.7 |
74 |
The resistance range of blaTEM-containing isolates varied from 82% to 100% against all tested â-lactam antibiotics. The level of resistance was appeared almost similar to those isolates without harboring the blaTEM gene. blaSHV-containing isolates showed almost equal levels of resistance compared to isolates without carrying blaSHV. All the 26 isolates carrying blaSHV showed 100% harmony of phenotypic resistance to cefixime, a third-generation cephalosporin. However, 93% of the isolates with blaSHV also showed resistance to the antibiotic. Association of blaOXA was somewhat stronger than those of blaTEM and blaSHV, incurring phenotypic resistance to â-lactam antibiotics. Isolates carrying blaOXA showed overall higher phenotypic resistance; notably, resistance to ceftriaxone was 61% in blaOXA-positive isolates that 40% in blaOXA-negative bacteria.
Altogether, there were no statistically significant associations found between the antimicrobial susceptibility and the presence of the ESBL genes. A smaller sample size may affect the statistical association observed in this study.
A total of 113 poultry gut bacteria was shown segmented in pie-chart according to a different genus and species level. The most prevalent isolate was Proteus spp. (62.5%), followed by Escherichia coli (20.2%) and Klebsiella spp (11.1%). Some less frequent isolates, namely Enterobacter, Kurthiapopuli, Cronobacter spp., and Eikenella corrodens were shown together in ‘other’ segment.
The comparative frequency of the three ESBL genes, blaTEM, blaOXA, and blaSHV has been shown in different enteric bacteria isolated under this study. Y- axis represents the percentage value of the ESBL genes in the respective bacteria shown in the X-axis. blaTEM was identified over 50% of all the different isolates. Some isolates revealed overlapping carriage of two or three genes simultaneously. However, overlapping fractions were not shown here. Identification of blaOXA and blaSHV genes varied species to species. All of the Pseudomonas spp., carry blaOXA and none of the Shigella spp., carry blaSHV.
Discussion
Poultry faeces is the source of diverse microorganisms. Besides gram-positive bacteria, a substantial number of pathogenic bacteria also reported in poultry feces.23 We reported the highest frequency of Proteus spp. in poultry feces examined under this study. The higher abundance of Proteus spp. in chicken gut content showed harmony with a similar previous report from Bangladesh.24 Unlikely, some other studies had identified E.coli and Klebsiella spp. as the most frequent isolates from poultry droppings; we identified these bacteria as the next most abundance isolates after Proteus spp.25-27 An endemic poultry bacteria, Salmonella, which can potentially contaminate fresh produce or the environment, was also detected in this study.28,29 This study has reported some bacteria for the first time in poultry gut-content from Bangladesh. Those uncommon bacteria include Kurthia populi, Cronobacter spp. and Eikenella corrodens.
The increase of antibiotic resistance and the existence of multidrug-resistant ESBL producers have become an emerging issue worldwide. The indiscriminate use of antimicrobials in animal farming is likely to accelerate the development of AMR in pathogens, as well as in commensal organisms. In the current study, all isolates’ antimicrobial susceptibility patterns reveal that all of the isolates were resistant against most of the antibiotics included in the study. Moreover, all of the Klebsiella spp., Shigella spp., and Pseudomonas spp., showed more resistance than other isolates. Further, none of the beta-lactam antibiotics was useful toward Pseudomonas spp. Some reports showed that E. coli has a higher propensity to develop resistance.30 However, resistant pattern observed against ceftriaxone to Proteus spp. and Klebsiella spp. appeared higher than E. coli in this study. This study indicated that the isolated bacteria showed a very high resistance towards most of the antibiotic tested. The resistance pattern of poultry isolates against â-lactam antibiotics was higher than the previous report of Bangladesh.31 Moreover, carbapenem was found to be effective in the previous report, but lower susceptibility was observed here.3 Findings indicate that the resistance phenomena against cutting-edge antibiotics have been increasing since the last decades.
The increasing rate of antimicrobial resistant bacteria is a global problem that affects both human and animal ecosystems. The study showed that highly ESBL gene producing bacteria were circulating in poultry feces. blaTEM was the most prevalent ESBL gene detected in this study, followed by blaOXA and blaSHV, and collectively the ESBL genes revealed higher than previously published studies from the poultry sector.32 Over the counter availability of antibiotics, no or less stringent rules about their application in agriculture and animal husbandry, and lack of educated human resources are some remarkable factors that contribute to applying irrational antibiotics in poultry flocks. The aberrant uses of antibiotics may eventually enforce acquisition and transmission of both phenotype and genotype AMR in poultry, human, and environment. Further detailed studies should be undertaken to investigate more about these ever-increasing genetic hazards. The bacteria-specific analysis detected a higher ratio of blaTEM in E.coli, supported by multiple previous studies.33, 34 Likely, blaSHV was detected higher in E.coli and Klebsiella spp. in this study and previously.35 It was reported that some poultry-borne E. coli functions as a potential reservoir of AMR genes that may be transmitted to humans conveniently.36
This study showed a significant portion of isolates resistant to many â-lactam antibiotics without carrying either of the three ESBL genes. This observation makes the blaTEM, blaOXA, and blaSHV genes as some unnecessary entities for contributing resistance to â-lactam antibiotics. The disagreement of the genotype-phenotype association could be explained by other ESBL genes or factors that have not been investigated in this study.37, 38 Therefore, there are about 200 different types of ESBL genes responsible for conferring resistance to â-lactam antibiotics.39 Varieties of ESBLs genes out of the blaTEM, blaOXA, and blaSHV lineage may have contributed to the different phenotypic resistance phenomena.40 Previous studies suggest that the distribution of ESBL genotypes can vary in different geographical locations.41 Therefore, studies and surveillance covering broad geographic regions of Bangladesh could validate our present findings of ESBL genes and associated AMR phenotypes.
This study carries several basic limitations. This study was performed in a cross-sectional assessment and follow up could not be carried out due to resource limitations. The convenience sampling was undertaken; however, samples were collected from several districts of Bangladesh to secure some generalizability. This study analyzed only a few ESBL genes and limited b-lactam antibiotics. The small sample size was also a limiting factor in performing fully powered statistical analyses. However, our results were generated from a resource-limited setting and maintained internal validity by repeating independent experiments where necessary.
Conclusion
ESBL gene producing bacteria have been increasingly recognized in the poultry sector in Bangladesh. The high prevalence of multidrug-resistant bacteria in poultry environments may increase the risk of spread to humans, particularly to those who work close to poultry farms and their excretory products. The high level of antibiotic resistance in poultry faeces from Bangladesh indicates the higher presence of ESBL genes that could impact adverse effects on human-, animal-, and environmental-health.
References
- Nikaido H. Multidrug resistance in bacteria. Annual review of biochemistry. 2009;78:119-46.
DOI:78.082907.145923 - O’neill J. Antimicrobial resistance. Tackling a Crisis for the Health and Wealth of Nations. 2014.
Available from:www.who.int/antimicrobial-resistance/news/amr-newsletter-no13-july2016.pdf - Akinbami OR, Olofinsae S, Ayeni FA. Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria. PeerJ.6:e5053.
DOI:10.7717/peerj.5053 - Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N. A review on antibiotic resistance: alarm bells are ringing. Cureus. 2017;9.
DOI:10.7759/cureus.1403 - Roe MT, Pillai SD. Monitoring and identifying antibiotic resistance mechanisms in bacteria. Poultry science.
DOI:10.1093/ps/82.4.622 - Gajamer VR, Bhattacharjee A, Paul D, Ingti B, Sarkar A, Kapil J, et al. Emergence of Multidrug-Resistant Uropathogens harboring ESBL, Carbapenem, Aminoglycosides and AmpC resistant genes from Northern India. bioRxiv.375501.
DOI:10.1101/375501 - Chantziaras I, Boyen F, Callens Bnd, Dewulf J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. Journal of antimicrobial chemotherapy.
DOI:10.1093/jac/dkt443 - Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental health perspectives.
DOI:10.1289/ehp.1206446 - Maciuca IE, Williams NJ, Tuchilus C, Dorneanu O, Guguianu E, Carp-Carare C, et al. High prevalence of Escherichia coli-producing CTX-M-15 extended-spectrum beta- lactamases in poultry and human clinical isolates in Romania. Microbial Drug Resistance.21:651-62.
DOI:10.1089/mdr.2014.0248 - van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics: links between animals and humans. International journal of antimicrobial agents.
DOI:10.1016/S0924-8579(00)00145-X - Klüsener B, Young JJ, Murata Y, Allen GJ, Mori IC, Hugouvieux V, et al. Convergence of calcium signaling pathways of pathogenic elicitors and abscisic acid in Arabidopsis guard cells. Plant physiology. 2002;130:2152-63.
DOI:10.1104/pp.012187 - Bush K, Bradford PA. beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb Perspect Med. 2016;6. Epub 2016/06/23.
DOI:10.1101/cshperspect.a025247 - Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos A-P, Mevius D, et al. Public health risks of enterobacterial isolates producing extended-spectrum â-lactamases or AmpC â-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clinical infectious diseases. 2013;56:1030-7.
DOI:10.1093/cid/cis1043 - Doi Y, Paterson D, Egea P, Pascual A, López-Cerero L, Navarro M, et al. Extended-spectrum and CMY-type b- lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clinical Microbiology and Infection. 2010;16:33-8.
DOI:10.1111/j.1469-0691.2009.03001.x - Fang G, Li W, Shen X, Perez-Aguilar JM, Chong Y, Gao X, et al. Differential Pd-nanocrystal facets demonstrate distinct antibacterial activity against Gram-positive and Gram- negative bacteria. Nature communications.9:1-9.
DOI:10.1038/s41467-017-02502-3 - Saliu E-M, Vahjen W, Zentek J. Types and prevalence of extended–spectrum beta–lactamase producing Enterobacteriaceae in poultry. Animal health research reviews. 2017;18:46-57.
DOI:10.1017/S1466252317 000020 - Boonyasiri A, Tangkoskul T, Seenama C, Saiyarin J, Tiengrim S, Thamlikitkul V. Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathogens and global health.108:235-45.
DOI:10.1179/2047773214Y.0000000148 - Tekiner ÝH, Özpýnar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. brazilian journal of microbiology. 2016;47:444-51.
DOI:10.1016/j.bjm.2015.11.034 - Nikolich MP, Hong G, Shoemaker NB, Salyers AA. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255-60.
PMID:7944364 - Cappuccino JG, Sherman N. Microbiology: a laboratory manual (Vol. 9). Pearson/ Benjamin Cummings; 2008.
DOI: - Wayne PA. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 29th Edition
Available from:clsi.org/media/2663/m100ed29_sample.pdf - Ahmed OB, Dablool AS. Quality improvement of the DNA extracted by boiling method in gram negative bacteria. International Journal of Bioassays. 2017; 6:5347-9.
DOI:10.21746/ijbio.2017.04.004 - Bolan NS, Szogi AA, Chuasavathi T, Seshadri B, Rothrock Jr MJ, Panneerselvam P. Uses and management of poultry litter. World’s Poultry Science Journal. 2010;66:673-98.
DOI:10.1017/S0043933910000656 - Nahar A, Siddiquee M, Nahar S, Anwar KS, Islam S. Multidrug resistant-proteus mirabilis isolated from chicken droppings in commercial poultry farms: Bio-security concern and emerging public health threat in Bangladesh. Journal of Biosafety & Health Education. 2014; 2:120.
DOI:10.4172/2332-0893.1000120 - Akond MA, Alam S, Hassan SMR, Shirin M. Antibiotic resistance of Escherichia coli isolated from poultry and poultry environment of Bangladesh. Internet Journal of Food Safety. 2009;11:19-23.
DOI: - Ogunleye AO, Okunlade AO, Jeminlehin FO, Ajuwape ATP. Antibiotic resistance in Escherichia coli isolated from healthy cattle at a major cattle market in Ibadan, Oyo State, South Western, Nigeria. African Journal of Microbiology Research.7:4572-5.
DOI:10.5897/AJMR2013.6028 - Kim HB, Park CH, Kim CJ, Kim E-C, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrobial agents and chemotherapy. 2009;53:639-45.
DOI:10.1128/AAC.01051-08 - Wilkinson KG, Tee E, Tomkins RB, Hepworth G, Premier R. Effect of heating and aging of poultry litter on the persistence of enteric bacteria. Poultry science. 2011;90:10-8.
DOI:10.3382/ps.2010-01023 - Chinivasagam HN, Redding M, Runge G, Blackall PJ. Presence and incidence of food-borne pathogens in Australian chicken litter. British poultry science.51:311-8.
DOI:10.1080/00071668.2010.499424 - Varga C, Rajiæ A, McFall ME, Reid-Smith RJ, Deckert AE, Pearl DL, et al. Comparison of antimicrobial resistance in generic Escherichia coli and Salmonella spp. cultured from identical fecal samples in finishing swine. Canadian Journal of Veterinary Research. 2008;72:181.
PMID:18505208 - Hasan B, Faruque R, Drobni M, Waldenström J, Sadique A, Ahmed KU, et al. High prevalence of antibiotic resistance in pathogenic Escherichia coli from large-and small-scale poultry farms in Bangladesh. Avian diseases. 2011;55:689-92.
DOI:10.1637/9686-021411-Reg.1 - Abreu R, Castro B, Espigares E, Rodríguez-Álvarez C, Lecuona M, Moreno E, et al. Prevalence of CTX-M-type extended-spectrum â-lactamases in Escherichia coli strains isolated in poultry farms. Foodborne pathogens and disease. 2014;11:868-73.
DOI:10.1089/fpd.2014.1796 - Khoshbakht R, Seifi S, Raeisi M. Antibiotic susceptibility and high prevalence of extended spectrum beta-lactamase producing Escherichia coli in iranian broilers. Revue de Medecine Veterinaire. 2016;167:133-7.
Available from:www.revmedvet.com/2016/RMV167_133_137.pdf - Osman KM, Kappell AD, Elhadidy M, ElMougy F, Abd El- Ghany WA, Orabi A, Mubarak AS, Dawoud TM, Hemeg HA, Moussa IM, Hessain AM. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Scientific reports. 2018;11;8:1-4.
DOI:10.1038/s41598-018-23962-7 - Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum â-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infection, Genetics and Evolution. 2011;11:1499-504.
DOI:10.1016/j.meegid.2011.06.001 - Nhung NT, Chansiripornchai N, Carrique-Mas JJ. Antimicrobial resistance in bacterial poultry pathogens: a review. Frontiers in veterinary science. 2017;10:126.
DOI:10.3389/fvets.2017.00126 - Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42. Epub 2017/10/27.
DOI:10.1093/femsre/fux053 - Bush K. Past and Present Perspectives on beta-Lactamases. Antimicrob Agents Chemother. 2018;62. Epub 2018/08/01.
DOI:10.1128/AAC.01076-18 - Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933-51, table of contents. Epub 2001/10/05.
DOI:10.1128/CMR.14.4.933-951.2001 - Paterson DL, Bonomo RA. Extended-spectrum beta- lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657-86. Epub 2005/10/15.
DOI:10.1128/CMR.18.4.657-686.2005 - Sheng WH, Badal RE, Hsueh PR. Distribution of extended- spectrum beta-lactamases, AmpC beta-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother. 2013;57:2981-8. Epub 2013/04/17.
DOI:10.1128/AAC.00971-12
Department of Microbiology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
salequl@juniv.edu
 0000-0001-6131-4132
0000-0001-6131-4132
Submission
19 July 2020
Accepted
30 June 2021
Published
01August 2021
Apply citation style format of Bangladesh Medical Research Council
Issue
Vol 47 No 2 (2021)
Section
Research Articles
Ethical Clearance
NREC of Bangladesh Medical Research Council (BMRC).
Financial Support
Bangladesh Medical Research Council (BMRC).
Conflict of Interest
None of the authors declared competing or conflict of interest.