Dr. Laila Khaleda
Department of Genetic Engineering and Biotechonology, University of Chittagong, Chattogram, Bangladesh.
Israt Akter Priya
Department of Genetic Engineering and Biotechonology, University of Chittagong, Chattogram, Bangladesh.
Md. Abdur Rahman Apu
Department of Genetic Engineering and Biotechonology, University of Chittagong, Chattogram, Bangladesh.
Amit Datta
Department of Genetic Engineering and Biotechonology, University of Chittagong, Chattogram, Bangladesh.
Basana Rani Muhuri
Department of Pediatric Nephrology, Chittagong Medical College Hospital, Chattogram, Bangladesh.
Mohammad Al-Forkan
Department of Genetic Engineering and Biotechonology, University of Chittagong, Chattogram, Bangladesh.
Keywords: Autism spectrum disorder, NLGN gene, Genetic abnormalities, Birth complications
DOI: 10.3329/bmrcb.v47i3.59237
Abstract
Background: Autism Spectrum Disorder (ASD) is a group of neurodevelopmental disorder which is now a hotbed worldwide. Being located on chromosome X and playing a vital role in synaptic transmission, NLGN-3 and 4X have been drawn the attention of many researchers as the most essential and functional candidate genes for ASD pathogenesis. However, there are many contradictory results considering their role in developing ASD in different populations.
Objective: This study was aimed to investigate the association of NLGN-3 and 4X genes along with non-genetic factors to develop ASD in Bangladeshi population.
Methods: In this study, we analysed rs4844285 and rs11795613 of NLGN-3 gene and rs3810686 and rs6638575 of NLGN-4X gene for both family-based association analysis and case-control based association analysis using polymerase chain reaction (PCR) and DNA sequencing. Along with this, demographic data were also analysed. A total of 60 members of 15 families, including ASD subjects and another 60 ASD subjects and 60 healthy people were included in this study.
Results: Allele A and genotype AA of NLGN-3 rs4844285 were found to be a probable risk factor for developing ASD in our studied population (p=0.031 for allele A, S c2= 2.707, OR=1.833 for genotype AA). We also found different birth complications in this studied population which can be considered to intensify the risk of ASD along with the abnormalities of genes.
Conclusion: From this study, it is clear that not only genetic abnormalities but also some birth anomalies play a vital role for the risk of developing ASD.
Keywords: Autism spectrum disorder, NLGN gene, Genetic abnormalities, Birth complications.
Introduction
Autism Spectrum Disorder (ASD) is a condition that appears very early in childhood development, varies in severity, and is characterised by a deficiency in acculturation, defect in lingual and gestural communications, and tiresome pattern of behavior.1,2 People with ASD may also experience sleeping problems and irritability. Sometimes they fell anxiety, over-responsivity, and gastrointestinal problems.3-5 Epilepsy, dental issues, and other mental problems are also found to be shared in some ASD patients.6 Both genetic and environmental aspects are thought to prompt in advancing the ASD risk, but the actual reason is still unknown.
The prevalence of ASD is about 1% worldwide, and in the larger picture, 1 in 59 children are affected with ASD.7,8 Two different population-based studies have found this disorder to be extremely heritable at a rate of about 40-80%.9,10 Study has revealed that monozygotic twins are at higher risk of developing ASD than dizygotic twins.11
More than 20% of the world’s population resides in the South Asia region, but the frequency of ASD occurrence is still mainly in the dark in this region.12 In Bangladesh, the reported prevalence was 0.2%, 0.84%, and 0.15%.13-15 According to the research findings by the Ministry of Social Welfare, Bangladesh
2016, 19% of total neurological disabilities was recorded as the rate of autism.16 Moreover, almost 3,00,000 children are suffering from ASD where 1 in every 150 girls and 1 in every 94 boys having ASD in Bangladesh according to Autistic Children’s Welfare Foundation, 2020.17
ASD susceptible genes have been identified by genome-wide linkage studies and genetic association studies, where most of which are involved in the development of the brain.18,19 As ASD is prevalent in male than female, the role of the X chromosome in the etiology of ASD has been under consideration, and thus, both NLGN-3 and NLGN-4X genes are thought as good positional and functional candidate genes in ASD susceptibility.20
Neuroligin a cell surface adhesion protein that is homologous to acetylcholinesterase and other esterases, binds to b-neurexin, a cell surface protein to form functional synapses.21,22 Neuroligins family includes five members of type 1 transmembrane protein of which NLGN3, NLGN4X, and NLGN4Y are located on sex chromosomes.23 Proteins encoded by NLGN genes are mostly expressed in the brain and interact with presynaptic NRXNs in a Calcium-dependent manner.24 The X-linked NLGN3 and NLGN4X are the first discovered ASD-associated genes in this pathway.25
Neuroligin 3 (NLGN3) gene, located on Chr: Xq13 and composed of eight exons with a start codon in exon 2.26 Two brothers with autism and Asperger syndrome respectively were first reported to have a point mutation on NLGN3 (R451C).25 NLGN4, the fourth member of the neuroligin family gene, is located on Chr: Xp22.3 with six exons.27 Study has found that 1bp insertion in the NLGN4 gene causes a frameshift mutation and thus premature termination (D396X), while another study revealed 2bp deletion causes nonspecific X-linked mental retardation in an individual with or without autism or PDD-NOS.25,28
In Bangladesh, number of cases of ASD is about 10.5 lakhs, however, unfortunately, this disorder is one of the least understood ones in our country.29 Comparing with the statistical data, research on a molecular level to assess the genetic basis of ASD is at the nascent stage in our country. This study thus aimed to identify the actual cause of ASD in this population and to quest if there is any connection between ASD and single nucleotide polymorphisms of NLGN-3 and NLGN-4X genes.
Materials and Methods
Study subjects and collection of samples: A total of 60 members of 15 families, including ASD children, were analysed in family-based association analysis. Another 60 ASD children and 60 healthy people without any previous history of neurological disorder were recruited for case-control based association analysis. The ASD patients were diagnosed with the diagnostic and statistical manual of mental disorder-IV criteria autistic disorder. The age of the ASD children was between 4-30 years. Three milliliter of peripheral blood was collected from each of the subjects. Questionnaire was provided to all the patients to obtain demographic data. Ethical clearance was taken from the Ethical Review Board (ERB) of Chittagong Medical College and Hospital, Chittagong. Each family was informed about this study and written informed consent was obtained from them.
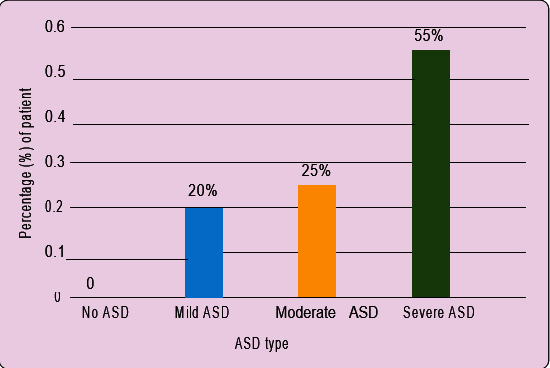
Categorisation of patients: Based on the ASD Assessment Scale/Screening Questionnaire, the patients were divided into four categories, i.e. 0-49=No ASD, 50-100=Mild ASD, 101-150=Moderate ASD, and >150=Severe ASD.
Selection of SNPs: Four SNPs of NLGN-3 and NLGN- 4X genes were selected based on the literature study.30, 31 The selected SNPs and their position in the chromosome were also recorded in (table I).
PCR amplification and sequencing: Genomic DNA was extracted from the whole blood sample using the standard phenol-chloroform method. The entire coding regions of NLGN-3 and 4X gene for all the subjects were analyzed by PCR and direct sequencing using four pairs of primers (table II). The cycling conditions consisted of 35 cycles with an initial denaturation at 95ºC for 5 minutes following a denaturation event of 30 seconds at 95ºC, annealing at 57ºC for 30 seconds, and finally an elongation period of 30 seconds at 72°C.
Statistical analysis: All the statistical analyses in this study were performed with the help of SPSS V.26. Both allelic and genotypic distribution for case and control samples besides with Odds ratio (OR) were set on using Fisher’s exact test. The two-sided test results were considered to be appropriate. p-value <0.05 was considered to be significant.
Results
ASD severity analysis: Based on ASD Assessment Scale/Screening, we found 33 patients at severe ASD stage and 15 and 12 patients at moderate and mild ASD states respectively (Figure 1).
|
Gene |
Gene Product |
SNPs |
Length (bp) |
Position of SNPs |
Alleles |
|---|---|---|---|---|---|
|
NLGN-3 |
Neuroligin- 3 |
rs11795613 |
124 |
chrX:7114748 |
T>C |
|
|
|
rs4844285 |
86 |
chrX:71150394 |
G>A |
|
NLGN-4X |
Neuroligin- 4x |
rs6638575 |
191 |
chrX5894600 |
A>G |
|
|
|
rs3810686 |
186 |
chrX:5892533 |
C>A/C>G/C>T |
|
SNPs |
Primers |
Reverse Primer |
|---|---|---|
|
Forward Primer |
||
|
NLGN-3 rs11795613 |
5'-CCTGGGCATGTGAAACCT-3' |
5'-CCTGTGGAAAGAGGGAAGTA-3' |
|
NLGN-3 rs4844285 |
5'-TCTGAGGTTGGTAGGGTACAGT-3' |
5'-CCTGCAGGTTTAAGAGACCTT-3' |
|
NLGN-4X rs6638575 |
5'-TTTTGAAGGGGAAGTGTTATC-3' |
5'-AGTCAAGGGCTGTTACAGAT-3' |
|
NLGN-4X rs3810686 |
5'-ACAAGATCAACTTCTGACCCT-3' |
5'-TATCAAGTGTCCTTGGCTGAG-3' |
Family-based association analysis: Evaluation of the sequencing data disclosed that all the 15 ASD patients have genotype AA (100%) for NLGN-3 rs4844285 while most of the parents and siblings possessed GG genotype (90% and 80% respectively). That means this genotype was not inherited from either parent. On the other hand, no significant association was found for rs11795613 of NLGN-3 gene with ASD in our studied population (table III).
|
Gene |
Genotype |
Number (%) of Patients(n=15) |
Number (%) of Parents(n=30) |
Number (%) of Siblings(n=15) |
|---|---|---|---|---|
|
NLGN-3rs4844285 |
AA |
15(100%) |
3(10%) |
3(20%) |
|
|
AG |
0 |
0 |
0 |
|
|
GG |
0 |
27(90%) |
12(80%) |
|
NLGN-3rs11795613 |
GG |
9(60%) |
12(40%) |
3(20%) |
|
|
AG |
0 |
6(20%) |
3(20%) |
|
|
AA |
6(40%) |
12(40%) |
9(60%) |
|
NLGN-4Xrs6638575 |
AA |
0 |
0 |
1(20%) |
|
|
AG |
0 |
9(30%) |
3(20%) |
|
|
GG |
15(100%) |
21(70%) |
9(60%) |
|
NLGN-4Xrs3810686 |
CC |
3(20%) |
12(40%) |
15(100%) |
|
|
CT |
3(20%) |
6(20%) |
0 |
|
|
TT |
9(60%) |
12(40%) |
0 |
For NLGN-4X rs6638575, it was observed that the GG genotype was frequent in patients (100%) as well as in their parents (70%) and siblings (60%). On the contrary, CC genotype of rs3810686 of NLGN-4X gene was dominant in siblings (100%) while most of the patients and their parents have TT genotype (60% and 40% accordingly) (table III).
|
GeneSNPs |
MAF (Case) |
MAF (Control) |
p Value |
c2 |
OR95% CI |
|---|---|---|---|---|---|
|
NLGN-3rs4844285(G>A) |
0.60 |
0.45 |
0.028 |
5.414 |
1.833(1.098-3.061) |
|
NLGN-3rs11795613(A>G) |
0.35 |
0.40 |
0.088 |
3.380 |
0.615(0.366-1.034) |
|
NLGN-4Xrs6638575(A>G) |
0.30 |
0.225 |
0.240 |
1.743 |
1.476(0.827-2.636) |
|
NLGN-4Xrs3810686 |
0.425 |
0.475 |
0.517 |
0.606 |
10.817(0.491-1.359) |
|
(C>A/C>G/C>T) |
|
|
|
|
|
Case-control based association analysis: A significant genetic association between NLGN-3 rs4844285 and ASD was observed from the case-control based analysis, with A allele to be considered as one of the risk factors of ASD (p=0.028, c2=5.414, OR=1.833, 95% CI=1.098-3.061). Both case and control samples displayed a similar type of genotypic and allelic pattern for the rest of the rs of the NLGN-3 gene and the two rs of NLGN-4X gene (table IV, table IA).
|
GeneSNP |
Genotype |
No. in cases |
No. in controls |
|---|---|---|---|
|
NLGN-3rs4844285 |
GG |
24 |
33 |
|
|
AA |
36 |
27 |
|
NLGN-3rs11795613 |
GG |
21 |
28 |
|
|
AA |
39 |
32 |
|
NLGN-4Xrs6638575 |
GG |
42 |
45 |
|
|
AG |
0 |
3 |
|
|
AA |
18 |
12 |
|
NLGN-4Xrs3810686 |
TT |
24 |
27 |
|
|
CC |
33 |
30 |
|
|
TC |
3 |
3 |
Analysis of demographic data: Interpretation of demographic data on patients disclosed that all the selected patients had experienced some sort of birth complications during pregnancy period and after birth. C-section delivery was pervasive among all the patients. Besides, most of the patients suffered from anemia, pneumonia, and hypoxia immediately after birth or the first few weeks of birth. Some patients also underwent neonatal exposure to viruses like CMV, Rotavirus, etc. After analysing the demographic data, it was emanated that asthma, gout, allergy, thyroid problems, and autoimmune disorders were recurrent in most of the mother and diabetics, high blood pressure and cardiac issues were found to be frequent in most father. All the demographic data were also recorded (table IIA).
|
Patient No. |
Complications of the mother during pregnancy and delivery |
Complications of father |
Complications of the patient immediately after delivery |
Level of ASD of patient |
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
Asthma, c-section delivery |
High bp |
|
Severe |
|
|||||
|
2 |
Exposer to CMV, c-section delivery, |
High bp, cardiac |
Pneumonia |
Severe |
|
|||||
|
|
hypoxia |
issues |
|
|
|
|||||
|
3 |
Gout, c-section delivery |
|
|
Moderate |
|
|||||
|
4 |
Allergy, Premature delivery |
High bp |
|
Moderate |
|
|||||
|
5 |
Exposer to Rotavirus, c-section delivery |
Allergy |
Diarrhea |
Severe |
|
|||||
|
6 |
Post-mature delivery |
|
|
Mild |
|
|||||
|
7 |
Asthma, Premature delivery, hypoxia |
High bp |
|
Moderate |
|
|||||
|
8 |
Gout, Exposer to CMV, c-section delivery |
Diabetics |
Pneumonia |
Severe |
|
|||||
|
9 |
Exposer to Rotavirus, c-section delivery |
High bp |
Diarrhea |
Severe |
|
|||||
|
10 |
|
Cardiac issues |
|
Moderate |
|
|||||
|
11 |
Gout, Post-mature delivery |
|
|
Severe |
|
|||||
|
12 |
Thyroid problems, Premature delivery |
|
|
Mild |
|
|||||
|
13 |
Exposer to CMV, c-section delivery, hypoxia |
|
Pneumonia |
Severe |
|
|||||
|
14 |
Autoimmune disorders, Asthma, c-section |
High bp, |
|
Severe |
|
|||||
|
|
delivery |
diabetics |
|
|
|
|||||
|
15 |
Exposer to CMV, Post-mature delivery |
High bp |
Pneumonia |
Moderate |
|
|||||
|
16 |
|
|
|
Mild |
|
|||||
|
17 |
Autoimmune disorders, Exposer to |
|
Diarrhea |
Severe |
|
|||||
|
|
Rotavirus, c-section delivery |
|
|
|
|
|||||
|
18 |
Exposer to CMV, c-section delivery |
Cardiac issues |
Pneumonia |
Severe |
|
|||||
|
19 |
Asthma, Premature delivery |
|
|
Mild |
|
|||||
|
20 |
Exposer to Rotavirus, c-section delivery |
High bp |
Diarrhea |
Severe |
|
|||||
|
21 |
Exposer to Rotavirus, c-section delivery |
Allergy |
Diarrhea |
Severe |
|
|||||
|
22 |
Post-mature delivery |
|
|
Mild |
|
|||||
|
23 |
Asthma, Premature delivery, hypoxia |
High bp |
|
Moderate |
|
|||||
|
24 |
Exposer to CMV, c-section delivery |
Cardiac issues |
Pneumonia |
Severe |
|
|||||
|
25 |
Exposer to CMV, c-section delivery, hypoxia |
|
Pneumonia |
Severe |
|
|||||
|
26 |
Exposer to Rotavirus, c-section delivery |
Allergy |
Diarrhea |
Severe |
|
|||||
|
27 |
|
|
|
Mild |
|
|||||
|
28 |
Exposer to CMV, c-section delivery, hypoxia |
|
Pneumonia |
Severe |
|
|||||
|
29 |
Exposer to Rotavirus, c-section delivery |
High bp |
Diarrhea |
Severe |
|
|||||
|
30 |
Thyroid problems, Premature delivery |
|
|
Mild |
|
|||||
|
31 |
Exposer to CMV, c-section delivery, hypoxia |
|
Pneumonia |
Severe |
|
|||||
|
32 |
|
Cardiac issues |
|
Moderate |
|
|||||
|
|
33 |
Gout, Post-mature delivery |
|
|
Severe |
|||||
|
|
34 |
|
|
|
Severe |
|||||
|
|
35 |
Post-mature delivery |
|
|
Mild |
|||||
|
|
36 |
Gout, c-section delivery |
|
|
Moderate |
|||||
|
|
37 |
|
|
|
Severe |
|||||
|
|
38 |
Asthma, Premature delivery, hypoxia |
High bp |
|
Moderate |
|||||
|
|
39 |
Premature delivery |
|
|
Severe |
|||||
|
|
40 |
|
|
|
Mild |
|||||
|
|
41 |
Exposer to Rotavirus, c-section delivery |
Allergy |
Diarrhea |
Severe |
|||||
|
|
42 |
Exposer to CMV, c-section delivery |
Cardiac issues |
Pneumonia |
Severe |
|||||
|
|
43 |
Asthma, Premature delivery, hypoxia |
High bp |
|
Moderate |
|||||
|
|
44 |
Gout, c-section delivery |
|
|
Moderate |
|||||
|
|
45 |
Asthma, c-section delivery |
High bp |
|
Severe |
|||||
|
|
46 |
Autoimmune disorders, Exposer to Rotavirus, |
|
Diarrhea |
Severe |
|||||
|
|
47 |
|
|
|
Mild |
|||||
|
|
48 |
Exposer to Rotavirus, c-section delivery |
Allergy |
Diarrhea |
Severe |
|||||
|
|
49 |
Exposer to Rotavirus, post mature delivery |
High bp |
Diarrhea |
Severe |
|||||
|
|
50 |
|
|
|
Moderate |
|||||
|
|
51 |
Asthma |
|
|
Severe |
|||||
|
|
52 |
Premature delivery |
|
|
Mild |
|||||
|
|
53 |
|
|
|
Severe |
|||||
|
|
54 |
|
High bp |
|
Severe |
|||||
|
|
55 |
Thyroid problems |
|
|
Moderate |
|||||
|
|
56 |
|
|
|
Severe |
|||||
|
|
57 |
|
Cardiac issues |
|
Moderate |
|||||
|
|
58 |
Gout, Post-mature delivery |
|
|
Severe |
|||||
|
|
59 |
|
|
|
Mild |
|||||
|
|
60 |
Gout, c-section delivery |
|
|
Moderate |
|||||
Association of total ASD score with NLGN-3 and NLGN-4X genes: Genotype pattern AA-AA of the selected rs of NLGN-3 and GG-TT of the selected rs of NLGN-4X gene were found to be associated with causing severe ASD in most of the cases. In a few cases, GG-TT pattern of NLGN-4x gene was also found to be associated with a moderate level of ASD. An interesting fact was observed from both family- based analysis and case-control based analysis that these patterns of genotype was also present in parents, healthy siblings, and also in healthy controls (table V).
|
Genotype of NLGN-3 with ASD score |
Number of Patients (%) |
Genotype of NLGN-4X |
Number of Patients (%) |
||||
|
rs4844285 |
rs11795613 |
ASDCategory |
Rs6638575 |
rs3810686 |
ASD Category |
||
|
AA |
GG |
Severe |
12(20%) |
AA |
CC |
Severe |
3(5%) |
|
AA |
AA |
Severe |
15(25%) |
AA |
TT |
Severe |
3(5%) |
|
GG |
AA |
Severe |
9(15%) |
GG |
TT |
Severe |
15(25%) |
|
AA |
GG |
Moderate |
6(10%) |
GG |
CC |
Severe |
9(15%) |
|
GG |
AA |
Moderate |
12(20%) |
GG |
CT |
Severe |
3(5%) |
|
AA |
GG |
Mild |
3(5%) |
AA |
CC |
Moderate |
9(15%) |
|
GG |
AA |
Mild |
3(5%) |
GG |
CC |
Moderate |
6(10%) |
|
|
|
|
|
GG |
TT |
Moderate |
6(10%) |
|
|
|
|
|
AA |
CC |
Mild |
3(5%) |
|
|
|
|
|
GG |
CC |
Mild |
3(5%) |
Discussion
The case-control based study generated a significant association between SNPs in NLGN-3 and ASD in our studied population, but no notable result was found for NLGN-4X polymorphisms. In addition to this, the family-based association analysis did not provide any strong association. In the case-control study, one SNP of NLGN-3 (rs4844285) was significantly associated with ASD at a 5% level of significance. The A allele of NLGN-3 rs4844285 was found as the risk allele of ASD in our studied sample. These results may suggest a potential role of NLGN-3 in ASD, which is relevant to the previous studies.21,30
In contrast to the case-control study, no significant evidence was provided by the family-based study up to the limit of appreciation. This could be due to the limited number of families with insufficient information. For NLGN-4X rs6638575 G allele and GG genotype and for rs3810686 T allele and TT genotype were observed to be preferentially transmitted to the affected individuals and thus, undoubtedly point towards the inheritance postulation.
Based on the role of the X chromosome to develop ASD, previous studies observed some controversial results.20,23,32-34 A study on the Chinese population also found no involvement of these SNPs of NLGN-3 and 4X in the development of ASD but other SNPs (rs3747333 and rs3747334) of NLGN-4X gene were discovered to be correlated with ASD in the Chinese Han population.20,23 On the other hand, missense mutations in the neuroligin-4 gene were found to be associated with autism susceptibility in the Bulgarian population and four novel synonymous substitutions were found in the Japanese ASD population.32,33 So, this can be assumed that the effect of polymorphisms of the neuroligin gene varies depending on sample size and ethnicity as well as ASD heterogeneity.
One of the most important parts of this study is the analysis of demographic data of patients. From the data of the patient’s history and the disease history of family members, we have found that obstacles that occurred during pregnancy and at the time of birth have resulted in severe ASD stage in most of the cases and also found to be related with some moderate to a mild level of ASD. These complications like misposition of the baby, c-section delivery, premature or post- mature delivery, hypoxia, etc. directly or indirectly in some points, affect different regions of the brain and thus may be hypothesized to be responsible for ASD development. We also found that the rate of patients affected with pneumonia immediately after birth or after a few months of birth was also frequent in our sample. Diarrhoea and anaemia were also spotted to be persistent in mother and also in patients which may be pointed as a major cause of the defect in the brain. Thus, the birth complications and prenatal exposure to different viruses may also be anticipated as a causal factor of ASD like the previous studies where birth complications and environmental exposure were found to be related with ASD.35,36
From the history of diseases of the family members, we noticed that patients with a mother or father who have high blood pressure, cardiac issues, diabetics, and allergy, scored moderate to severe levels of ASD in our study population. Therefore, parental disorders may also be predicted to associate with ASD risk in our studied population along with genotypic dysfunction.
Most of the people of this region neglect necessary precautions during pregnancy which results in different complications at the time of labour and after the birth of the child. At this point, we may hypothesise that these complications are also becoming a risk factor for developing ASD in our population. Therefore, we can draw an end line by pointing on the fact that, these anomalies with the partial effect of a defect in selected genes may take to the court for intensifying the risk of inaugurating ASD in the current study population.
Conclusion
Polymorphisms in the NLGN-3 rs4844285 gene along with complications during pregnancy period and labour conditions are associated with developing ASD risk in our studied population. We did not find any noteworthy results for other SNPs for both family- based and case-control based study. This is because of the small sample size in our study. So, further study with larger sample size and more information about family members and patients may be recommended for exploring the association more accurately.
Acknowledgments
This study was supported by the grant of Bangladesh Medical Research Council (BMRC) and the grant of Advanced Research in Education (GARE), Ministry of Education, Bangladesh. The authors gratefully acknowledge the Nishpap Autism Foundation, Chittagong, and the Prerona Autism School, Chittagong for their committed support and encouragement in the management of patients and parents. The authors also like to extend their gratitude to Mrs Rawshan Jahan (Teacher, Nishpap Autism Foundation) for her helping hand in sample collection.
References
- Jacob S, Brune CW, Carter CS, Leventhal BL, Lord C, Cook Jr EH. Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neuroscience letters. 2007; 417:6-9.
Doi:10.1016/j.neulet.2007.02.001 - Liu X, Kawamura Y, Shimada T, Otowa T, Koishi S, Sugiyama T, Nishida H, Hashimoto O, Nakagami R, Tochigi M, Umekage T. Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. Journal of human genetics. 2010; 55:137-41.
Doi:org/10.1038/jhg.2009.140 - Mazurek MO, Vasa RA, Kalb LG, Kanne SM, Rosenberg D, Keefer A, Murray DS, Freedman B, Lowery LA. Anxiety, sensory over-responsivity, and gastrointestinal problems in children with autism spectrum disorders. Journal of Abnormal Child Psychology. 2013; 41:165-76.
Doi:10.1007/s10802-012-9668-x - Rudy BM, Lewin AB, Storch EA. Managing anxiety comorbidity in youth with autism spectrum disorders. Neuropsychiatry. 2013; 3:411.
Doi:10.2217/npy.13.53 - Molloy CA, Manning-Courtney P. Prevalence of chronic gastrointestinal symptoms in children with autism and autistic spectrum disorders. Autism. 2003 ; 7:165-71.
Doi:10.1177/1362361303007002004 - Ahmed SP, Ahmed MS. Risk of inter-related health issues among children with ASD in Bangladesh. Global Journal of Medical Research. 2015 13; 15:1-K.
Doi: - Safari MR, Omrani MD, Noroozi R, Sayad A, Sarrafzadeh S, Komaki A, Manjili FA, Mazdeh M, Ghaleiha A, Taheri M. Synaptosome-Associated Protein 25 (SNAP25) Gene Association Analysis Revealed Risk Variants for ASD, in Iranian Population. Journal of Molecular Neuroscience. 2017; 61:305.
Doi:10.1007/s12031-016-0860-2 - Autism and Developmental Disabilities Monitoring (ADDM) Network. Centers for Disease Control and Prevention. Centers for Disease Control and Prevention. 2019.
Available from:www.cdc.gov/ncbddd/autism/addm.html - Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, Metzger LM, Shoushtari CS, Splinter R, Reich W. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. Journal of Autism and Developmental Disorders. 2003;33: 427-33.
Doi:10.1023/A:1025014929212 - Ronald A, Happé F, Bolton P, Butcher LM, Price TS, Wheelwright S, Baron-Cohen S, Plomin R. Genetic heterogeneity between the three components of the autism spectrum: a twin study. Journal of the American Academy of Child & Adolescent Psychiatry. 2006;45: 691-99.
Doi:10.1097/01.chi.0000215325.13058.9d - Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. American Journal of Psychiatry. 2010; 167:1357-63.
Doi:10.1176/appi.ajp.2010.10020223 - Hossain MD, Ahmed HU, Uddin MJ, Chowdhury WA, Iqbal MS, Kabir RI, Chowdhury IA, Aftab A, Datta PG, Rabbani G, Hossain SW. Autism Spectrum Disorders (ASD) in South Asia: a systematic review. BMC Psychiatry. 2017 ; 17:1-7.
Doi:10.1186/s12888-017-1440-x - Mullick MS, Goodman R. The prevalence of psychiatric disorders among 5–10 years old in rural, urban and slum areas in Bangladesh. Social Psychiatry and Psychiatric Epidemiology. 2005 ; 40:663-71.
Doi:10.1007/s00127-005-0939-45 - Rabbani MG, Alam MF, Ahmed HU, Sarker M, Chowdhury WA, Zaman MM. Prevalence of mental disorders, mental retardation, epilepsy and substance abuse in children: A community based epidemiological survey. Bangladesh Journal of Psychiatry. 2009; 23:11-53.
Doi: - NCDC, RCHCIB, BMRC et al. Survey of Autism and Neurodevelopmental Disorders in Bangladesh, 2013. Non- communicable Diseases Control (NCDC) Programme, DGHS, MOHFW, Revitalization of Community Health Care Initiatives in Bangladesh (RCHCIB), Ministry of Health and Family Welfare (MOHFW), Bangladesh Medical Research Council (BMRC), MOHFW; Department of Pediatric Neuroscience, Dhaka Shishu Hospital, Dhaka, Bangladesh.
Doi: - Ministry of Social Welfare of Bangladesh. Disabilities Screening Bulletin. 2016; 27.
Available from:msw.gov.bd - Frequency of Autism. Autistic children’s Welfare Foundation, Bangladesh. 2020.
Available from:www.acwf-bd.org/frequency_autism.php - Gupta AR, State MW. Recent advances in the genetics of autism. Biological psychiatry. 2007 ; 61:429-37.
Doi:10.1016/j.biopsych.2006.06.020 - Vorstman JA, Staal WG, Van Daalen E, Van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Molecular Psychiatry. 2006 ; 11:18.
Doi:10.1038/sj.mp.4001757 - Landini M, Merelli I, Raggi ME, Galluccio N, Ciceri F, Bonfanti A, Camposeo S, Massagli A, Villa L, Salvi E, Cusi D. Association analysis of noncoding variants in neuroligins 3 and 4X genes with autism spectrum disorder in an Italian cohort. International Journal of Molecular Sciences. 2016; 17:1765.
Doi:10.3390/ijms17101765 - Zhao Y, Yang C, Lin H, Liang Y, Wang W, Chen H, Wang M, Liu H, Luo S, Yi Y. The single nucleotide polymorphism study on the SHANK3 and NLGN3 gene in association with autism in Wenzhou children. Int J Clin Exp Pathol. 2016 ; 9:5694-99.
Doi:www.ijcep.com/files/ijcep0018814 - Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005; 307:1324-28.
Doi:10.1126/science.1107470 - Xu X, Xiong Z, Zhang L, Liu Y, Lu L, Peng Y, Guo H, Zhao J, Xia K, Hu Z. Variations analysis of NLGN3 and NLGN4X gene in Chinese autism patients. Molecular Biology Reports. 2014; 41:4133-40.
Doi:10.1007/s11033-014-3284-5 - Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008 ; 455:903.
Doi:10.1038/nature07456 - Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genetics. 2003 ; 34:27.
Doi:10.1038/ng1136 - Philibert RA, Winfield SL, Sandhu HK, Martin BM, Ginns EI. The structure and expression of the human neuroligin-3 gene. Gene. 2000;246:303-10.
Doi:10.1016/S0378-1119(00)00049-4 - Bolliger MF, Karl FR, Kaspar H, GLOOR SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochemical Journal. 2001 ; 356:581-8.
Doi:10.1042/bj3560581 - Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neur oligin family. The American Journal of Human Genetics. 2004 ; 74:552-57.
Doi:10.1086/382137 - Rahman MM. Autism spectrum disorders. Journal of Bangladesh College of Physicians and Surgeons. 2010; 28:143-4.
Doi:10.3329/jbcps.v28i3.6506 - Yu J, He X, Yao D, Li Z, Li H, Zhao Z. A sex-specific association of common variants of neuroligin genes (NLGN3 and NLGN4X) with autism spectrum disorders in a Chinese Han cohort. Behavioral and Brain Functions. 2011;7:13.
Doi:10.1186/1744-9081-7-13 - Qi H, Xing L, Zhang K, Gao X, Zheng Z, Huang S, Guo Y, Zhang F. Positive association of neuroligin-4 gene with nonspecific mental retardation in the Qinba Mountains Region of China. Psychiatric genetics. 2009 ; 19:1-5.
Doi:10.1097/YPG.0b013e3283088e54 - Yanagi K, Kaname T, Wakui K, Hashimoto O, Fukushima Y, Naritomi K. Identification of four novel synonymous substitutions in the x-linked genes neuroligin 3 and neuroligin 4X in japanese patients with autistic spectrum disorder. Autism research and treatment. 2012.
Doi:10.1155/2012/724072 - Avdjieva-Tzavella DM, Todorov TP, Todorova AP, Kirov AV, Hadjidekova SP, Rukova BB, Litvinenko IO, Hristova- Naydenova DN, Tincheva RS, Toncheva DI. Analysis of the genes encoding neuroligins NLGN3 and NLGN4 in Bulgarian patients with autism. Genet Couns. 2012 ; 23:505-11.
Doi: - Wermter AK, Kamp Becker I, Strauch K, Schulte Körne G, Remschmidt H. No evidence for involvement of genetic variants in the X linked neuroligin genes NLGN3 and NLGN4X in probands with autism spectrum disorder on high functioning level. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics.2008 ; 147:535-7.
Doi:10.1002/ajmg.b.30618 - Matson ML, Matson JL, Beighley JS. Comorbidity of physical and motor problems in children with autism. Research in Developmental Disabilities. 2011 ; 32: 2304-08.
Doi:10.1016/j.ridd.2011.07.036 - Diaz-Anzaldua A, Diaz-Martinez A. Genetic, environmental, and epigenetic contribution to the susceptibility to autism spectrum disorders. Revista de Neurologia. 2013 ; 57: 556- 568.
PMID:24288105
Department of Genetic Engineering and Biotechonology, University of Chattogram, Chattagong-4331, Bangladesh.
lkankhi@gmail.com
 0000-0002-1331-5875
0000-0002-1331-5875
Submission
21 December 2020
Accepted
17 October 2021
Published
01 December 2021
Apply citation style format of Bangladesh Medical Research Council
Issue
Vol 47 No 3 (2021)
Section
Research Articles
Financial Support
Bangladesh Medical Research Council (BMRC) and Advanced Research in Education (GARE), Ministry of Education, Bangladesh
Conflict of Interest
None.